Senecavirus A (SVA) is a single-stranded, positive sense, nonenveloped RNA virus classified as the only species in the genus Senecavirus within the Picornaviridae family.1 The SVA genome is approximately 7300 bases in length containing an open reading frame encoding a polyprotein of 2181 amino acids, encoding 12 proteins2: L-VP4-VP2-VP3- VP1-2A-2B-2C-3A-3B-3C-3D. The viral genome has the viral genome-linked protein at its 5’ end; the 5’ untranslated region contains a type IV internal ribosome entry site, and the 3’ untranslated region end is polyadenylated.1
An SVA infection can cause porcine idiopathic vesicular disease (PIVD), characterized by coronary band hyperemia, lameness, erosions, and vesicles in the oral skin mucosa, snout, interdigital space, and along the coronary bands.2 Lesions are clinically indistinguishable from other vesicular diseases including foot-and-mouth disease (FMD), vesicular stomatitis (VS), and vesicular exanthema of swine (VES). Based on previous studies of experimental infections, lesions appear between 3 to 5 days post inoculation (dpi) and resolve by day 14 to 21 dpi.3 A survey by Fernandes et al4 comparing pathogenicity of the historical SVA strain SVV-001 to the contemporary SVA strain SD15-26 showed that the historic isolate presents low virulence in finishing pigs. In contrast, a similar study conducted by Buckley et al5 showed growing pigs developed vesicular lesions when inoculated with either a historical or contemporary SVA isolate despite the difference in infection kinetics. However, these contrasting results of clinical presentation in pigs after infection with historical or contemporary SVA strains indicate that additional research is required to better understand host and viral factors involved in the pathogenicity of SVA strains.
Senecavirus A was first isolated by the US Department of Agriculture National Veterinary Service Laboratory in 1988 from stillborn piglets and piglets showing diarrhea.6 Subsequently, SVA was detected in 2002 as an adventitious virus in cell line culture PER.C6. This first viral isolate was named SVV-001, and the complete genome sequence was published in 2008.7 Since the initial detection, subsequent analyses of samples from pigs with vesicular disease-like clinical signs revealed the circulation of SVA in different regions worldwide.
Senecavirus A has been present in US swine populations since 1988, with a significant increase in cases in late 2014.6 In addition, SVA was isolated from sows displaying PIVD-like clinical signs in 2007 in Canada.8 Furthermore, vesicular disease outbreaks caused by SVA were reported in 2015 in the United States, Brazil, and China.9-11 The SVA detected in China was similar to SVA described in the United States and Brazil; however, after progressive dissemination among several Chinese provinces, current SVA strains were segregated into at least 5 phylogenetic clades.12 In early 2016, SVA-related vesicular disease was described in swine from Colombia, where the detected strains shared homology with previously recognized SVA strains in the United States.13 In the same year, Thailand reported the detection of SVA, which was closely related to the SVA strain initially identified in Canada.14 Vietnam detected SVA isolates in 2018 with high genetic identity with SVA strains from China.15 Recently, SVA was confirmed to be the cause of vesicular disease in swine from Chile, allegedly originating from SVA strains circulating in the United States.16
In Mexico, SVA is considered an exotic virus by the National Secretary of Agriculture and Rural Development; thus, disease caused by SVA mandates immediate notification to the National System of Epidemiological Surveillance.17 No official information or scientific reports of SVA in the swine population within the Mexican national territory have been previously reported. In this case report, we describe the genetic characterization of SVA collected from 2 related clinical cases of SVA infection in pigs from 2 regions in Mexico in 2021.
Animal care and use
This study was conducted at the Mexico-United States Commission for Prevention of Foot-and-Mouth Disease and Other Exotic Animal Diseases (CPA) according to good production practices in the pig farm manual implemented by the Ministry of Agriculture and Rural Development.
Case descriptions
Case 1
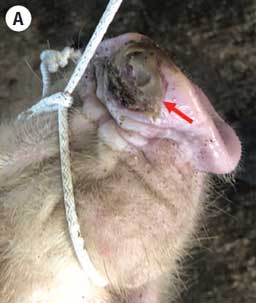
On December 21, 2021, a farm in the Valladolid municipality in Yucatán reported the sudden onset of vesicular lesions in 30 gilts at 20 weeks of age. These 30 gilts belonged to a 365-head group of replacement gilts ranging in age from 7 to 20 weeks old and were obtained from a farm in the state of Sonora in northern Mexico. Before the clinical manifestations, the gilts were transported and placed in quarantine at the destination farm in Yucatán. During arrival, lameness and fever were detected warranting further examination. Vesicles on the snouts and ulcerative lesions in the dewclaw, sole, heel, and coronary bands were detected (Figure 1). Sick animals were removed upon the onset of clinical signs and isolated from the quarantined gilts in separate pens off-site. Previous evidence of similar clinical manifestations on the farm or in neighboring areas was not reported. Due to the vesicular lesions resembling those of vesicular exotic diseases, drag swabs from the vesicular lesions of 6 affected animals and 1 epithelium tissue sample from the gilt with severe lesions were collected and submitted for diagnosis to the Immunology, Cellular, and Molecular Biology Laboratory from the CPA and identified as case number CPA-21738-21 (case 1).


Case 2
On December 22, 2021, the farm where the gilts from case 1 were obtained in Sonora state, reported ulcerative lesions on the snouts and mouths of 13 pigs. Due to the trade relationship and similar clinical manifestations with case 1, drag swabs, whole blood, and serum samples were collected from the 13 pigs with lesions and 2 epithelium samples were collected from pigs displaying severe lesions. Additionally, 2 whole blood and 2 serum samples were collected from clinically healthy pigs. All samples were submitted for diagnosis to the Immunology, Cellular, and Molecular Biology Laboratory from CPA under the case number CPA-21748-21 (case 2). Prior to the disease event, no evidence of vesicular lesions in the animal population was reported on this or neighboring farms.
Diagnosis and laboratory findings
Conventional differential diagnosis was completed by CPA to exclude diseases with related clinical presentations. The samples from case 1 were evaluated by real-time quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) for molecular screening of viruses associated with vesicular diseases including foot-and-mouth disease virus (FMDV), swine vesicular disease virus (SVDV), vesicular stomatitis virus (VSV), and SVA. No positive results were obtained for FMDV, SVDV, and VSV. Conversely, SVA was detected in 4 drag swab samples and 1 epithelium sample displaying positive cycle threshold values ranging from 22 to 33. In addition, viral isolation of SVA was attempted; however, no positive results were obtained.
Likewise, the qRT-PCR assay was used for FMDV, SVDV, VSV, and SVA detection in samples from case 2. Results indicated 2 epithelium and 1 whole blood sample were positive for SVA with positive cycle threshold values ranging from 28 to 34. Moreover, a competitive enzyme-linked immunosorbent assay was performed, and SVA antibodies were detected in all serum samples from case 2. In addition, a neutralization test and viral isolation for FMDV and VSV were performed with no positive results.
Genome sequencing of SVA detected in epithelium samples from both cases was conducted at the National Center for Diagnostic Services in Animal Health. The 2 SVA sequences, partial and whole genome, were deposited in GenBank under the accession numbers ON369393 and ON369394, respectively. The phylogeny based on the comparison of whole genome sequences of reference SVA strains has led to the segregation of SVA sequences into 3 main clades represented by prototype strains, historical strains, and contemporary strains. Similar to previous studies,18 there is a display of temporal and geographic clustering of SVA sequences. The clade of contemporary strains is particularly diverse as it includes circulating SVA identified between 2011 and 2021 and grouped by country of origin including strains from the United States, Canada, Colombia, Vietnam, China, Brazil, Thailand, and Mexico. The phylogenetic analysis clustered the SVA sequences from Mexico within the contemporary strains detected in the United States in 2017 (Figure 2). Furthermore, the genetic analysis revealed that SVA sequences from cases 1 and 2 presented a 99.9% nucleotide identity to each other and displayed a 92.7% identity with the SVV-01 SVA prototype strain and 98.2% with SVA strains detected in pigs from Canada and the United States. Specifically, based on the full-length genome sequence, the SVA identified in Mexico shared a 98.3% to 98.4% nucleotide identity and a 99.3% to 99.4% similarity at the amino acid level with an SVA detected in 2017 from swine in California named USA/MI17-011956/2017 (GenBank accession number MN812959.1).
The SVA amino acid sequences were deduced and analyzed for the presence of substitutions. Despite sharing a high genetic similarity, the amino acid substitution analysis showed differences between the SVA sequences from case 1 and case 2, revealed by 12 and 14 amino acid substitutions in the polyprotein sequence compared with the USA/MI17-011956/2017 strain, respectively (Table 1). The two Mexican SVA sequences exhibited more substitutions in the 3D gene, with 3 and 4 substitutions, respectively, followed by VP1, 2B, and 3A genes. Conversely, no mutations were found in L, VP4, VP2, 2A, and 3B genes.
| Gene | Range | Case 1 | Case 2 | |||
|---|---|---|---|---|---|---|
| Substitutions, No. | Amino acids | Substitutions, No. | Amino acids | |||
| L | 1-79 | 0 | – | 0 | – | |
| VP4 | 80-150 | 0 | – | 0 | – | |
| VP2 | 151-434 | 0 | – | 0 | – | |
| VP3 | 435-673 | 1 | Q492R | 1 | Q492R | |
| VP1 | 674-937 | 2 | V766A, I894V | 2 | V766A, I894V | |
| 2A | 938-946 | 0 | . | 0 | . | |
| 2B | 947-1074 | 2 | S1043R, K1060S | 2 | S1043R, K1060S | |
| 2C | 1075-1396 | 1 | T1317A | 1 | T1317A | |
| 3A | 1397-1486 | 2 | T1469A, E1472D | 2 | T1469A, E1472D | |
| 3B | 1487-1508 | 0 | . | 0 | . | |
| 3C | 1509-1719 | 1 | K1610R | 2 | A1519V, K1610R | |
| 3D | 1720-2180 | 3 | D1767G, A1850V, M1860V | 4 | D1767G, A1850V, M1860V, V1874A | |
Due to positive cases of SVA, both farms were quarantined, and control measures were conducted, including depopulation, farm disinfection, and surveillance.
Discussion
Since late 2014, a global increase in the occurrence of SVA infection cases in the United States, Brazil, Colombia, China, Thailand, Vietnam, and Chile has been reported.11-16 Phylogenetic analyses has shown the evolutionary divergence between SVA historical and contemporary strains and most SVA strains are grouped in separate phylogenetic clusters based on their geographic location. With a few exceptions, most contemporary clustered SVA strains are evolving independently within the swine population of an affected country. Therefore, transmission among nations can be inferred.19 Analysis of global SVA genomes revealed 3 major evolutionary clusters based on complete genome sequences or the VP1 gene sequence used. Hence, further investigation is needed for a definitive designation on SVA isolates emerging worldwide over time.20, 21
Mexico has been considered free of FMD since 1955, and other vesicular diseases like VS and VES are considered exotic diseases. Therefore, it is mandatory to rule out infectious diseases with clinical signs resembling these diseases. In late 2021, the detection of SVA was confirmed in 2 pig farms from northern and southwest Mexico, which were linked by pig movements. The affected animals displayed lameness, fever, vesicles on the snout, and ulcerative lesions in the interdigital region and coronary band. Senecavirus A was identified in different sample types including drag swabs from vesicular lesions, epithelium, and whole blood samples from the affected animals in case 1. Antibodies against SVA were detected in all serum samples collected from animals from case 2. In addition, the presence of other exotic vesicular diseases was ruled out.
In the present study, 1 full-length genome and 1 near-full-length genome (6071bp) of Mexican SVA, identified as CPA-21738-21 (case 1) and CPA-21748-21 (case 2), were obtained. Genetic characterization was performed to determine the phylogenetic relationship with previously reported SVA. Analysis of the genome sequences from both cases demonstrated that the Mexican SVA shared high genomic identity with each other (99.9%) at the nucleotide and amino acid levels. These SVA strains were grouped into the contemporary SVA strain cluster sharing a common ancestor with the 2017 and 2020 isolates from the United States because of the high genetic identity and similarity (Figure 2). Furthermore, the close genetic relationship between cases 1 and 2 suggests that they originated from the same ancestor but underwent different substitutions. Hence, SVA strains from Mexico have endured genetic changes in several regions including amino acid substitutions occurring in the 3D, VP1, and 3A genes compared with the USA/MI17-011956/2017 strain.
Similar to the SVA strains we identified, Bennett et al16 described that the SVA detected in Chile showed a close relationship with SVA detected in swine from California in 2017. Furthermore, the low homology in genetic identity of the SVA strains from Mexico in comparison with SVA strains from Colombia (96.9%), Brazil (95.7%), Vietnam (95.5%), Canada (95.2%), China (94.9%-95.1%), and Thailand (93.7%) demonstrated the isolates are distantly related.
In Mexico, SVA is classified as an exotic pathogen; therefore, outbreaks of vesicular disease caused by SVA have not been reported. Moreover, no SVA serosurveys have been conducted in the past. Nonetheless, SVA antibodies were detected in pigs with clinical signs from case 2. This suggests that SVA could have been circulating previously in the farm’s pig population for some period but was undetected due to the lack of detectable clinical presentation. Previous studies have shown that pigs infected with SVA might appear clinically healthy and remain asymptomatic.22,23 However, the development of vesicular lesions can be associated with immunosuppressive factors, like stress. Thus, activities like mobilization and transportation could potentially trigger the clinical presentation in SVA-infected animals.23 Likewise, the differences in nucleotide and amino acid levels between SVA strains identified in this study suggest continuous evolutionary events, possibly while discreetly spreading in the Mexican swine population before this first detection.
In countries with exotic vesicular diseases, finding SVA in swine has important implications and can lead to confusion in differentiating SVA from an exotic disease outbreak because they share similar clinical signs. Therefore, accurate SVA diagnosis by molecular screening and confirmation using serological assays are suggested as a more effective diagnostic method.11 Here, we have described the first detection of SVA in Mexico. The affected pigs were infected with a unique SVA isolate described herein, sharing homology with those SVA isolates identified in the United States in 2017.
Nonetheless, further studies of more cases need to be conducted to understand the source of SVA’s introduction to Mexico, its risk factors, prevalence, or detection of possible future outbreaks. Moreover, it is necessary to evaluate the transmission routes within pig populations and through mechanical vectors like flies. 24 In addition, retrospective serological studies from symptomatic and asymptomatic pigs will help determine the timing of SVA introduction in Mexico. These results will increase the knowledge regarding SVA epidemiology and highlight the significant SVA surveillance role in Mexican swine populations to prevent SVA reintroduction and further spread. Although control measures were applied in affected farms, SVA is now a growing concern for swine producers from Mexico. Thus, this case report will increase awareness that the prompt notification of the vesicular disease caused by SVA helps prevent and control SVA infection.
Implications
Under the conditions of this study:
- Evidence of SVA infection in pigs from Mexico was detected for the first time.
- Lesions and clinical signs of SVA can be misleading for diagnosis.
- Introduction of SVA in Mexico is a high-risk factor for swine producers.
Acknowledgments
Conflict of interest
None reported.
Disclaimer
Scientific manuscripts published in the Journal of Swine Health and Production are peer reviewed. However, information on medications, feed, and management techniques may be specific to the research or commercial situation presented in the manuscript. It is the responsibility of the reader to use information responsibly and in accordance with the rules and regulations governing research or the practice of veterinary medicine in their country or region.
References
1. Zell R, Delwart E, Gorbalenya AE, Hovi T, King AMQ, Knowles NJ, Lindberg AM, Pallansch MA, Palmenberg AC, Reuter G, Simmonds P, Skern T, Stanway G, Yamashita T, ICTV Report Consortium. ICTV virus taxonomy profile: Picornaviridae. J Gen Virol. 2017;98:2421-2422. https://doi.org/10.1099/jgv.0.000911
2. Cameron R. Diseases of skin. In: Straw BE, Zimmerman JJ, D’Allaire S, Taylor DJ, eds. Diseases of Swine. 9th ed. Blackwell; 2006:179-198.
3. Joshi LR, Fernandes MHV, Clement T, Lawson S, Pillatzki A, Resende TP, Vannucci FA, Kutish GF, Nelson EA, Diel DG. Pathogenesis of Senecavirus A infection in finishing pigs. J Gen Virol. 2016;97(12):3267-3279. https://doi.org/10.1099/jgv.0.000631
4. Fernandes MHV, Maggioli MF, Joshi LR, Clement T, Faccin TC, Rauh R, Bauermann FV, Diel DG. Pathogenicity and cross-reactive immune responses of a historical and a contemporary Senecavirus A strains in pigs. Virology. 2018;522:147-157. https://doi.org/10.1016/j.virol.2018.06.003
5. Buckley AC, Michael DD, Faaberg KS, Guo B, Yoon K-J, Lager KM. Comparison of historical and contemporary isolates of Senecavirus A. Vet Microbiol. 2021;253:108946. https://doi.org/10.1016/j.vetmic.2020.108946
*6. Knowles NJ, Hales LM, Jones BH, Landgraf JG, House JA, Skele KL, Burroughs KD, Hallenbeck PL. Epidemiology of Seneca Valley virus: Identification and characterization of isolates from pigs in the United States. In: XIVth Meeting of the European Study Group on the Molecular Biology of Picornaviruses. 2006;G2.
7. Hales LM, Knowles NJ, Reddy PS, Xu L, Hay C, Hallenbeck PL. Complete genome sequence analysis of Seneca Valley virus-001, a novel oncolytic picornavirus. J Gen Virol. 2008;89(Pt 5):1265-1275. https://doi.org/10.1099/vir.0.83570-0
8. Pasma T, Davidson S, Shaw SL. Idiopathic vesicular disease in swine in Manitoba. Can Vet J. 2008;49:84–85.
9. Vannucci FA, Linhares DC, Barcellos DE, Lam HC, Collins J, Marthaler D. Identification and complete genome of Seneca Valley virus in vesicular fluid and sera of pigs affected with idiopathic vesicular disease, Brazil. Transbound Emerg Dis. 2015;62(6):589-593. https://doi.org/10.1111/tbed.12410
10. Wu Q, Zhao X, Chen Y, He X, Zhang G, Ma J. Complete genome sequence of Seneca Valley virus CH-01-2015 identified in China. Genome Announc. 2016;4(1):e01509-15. https://doi.org/10.1128/genomeA.01509-15
11. Gimenez-Lirola LG, Rademacher C, Linhares D, Harmon K, Rotolo M, Sun Y, Baum DH, Zimmerman J, Piñeyro P. Serological and molecular detection of Senecavirus A associated with an outbreak of swine idiopathic vesicular disease and neonatal mortality. J Clin Microbiol. 2016;54(8):2082-2089. https://doi.org/10.1128/JCM.00710-16
12. Wang M, Chen L, Pan S, Mou C, Shi K, Chen Z. Molecular evolution and characterization of novel Seneca Valley virus (SVV) strains in South China. Infect Genet Evol. 2019;69:1-7. https://doi.org/10.1016/j.meegid.2019.01.004
13. Sun D, Vannucci F, Knutson TP, Corzo C, Marthaler DG. Emergence and whole-genome sequence of Senecavirus A in Colombia. Transbound Emerg Dis. 2017;64(5):1346-1349. https://doi.org/10.1111/tbed.12669
14. Saeng-Chuto K, Rodtian P, Temeeyasen G, Wegner M, Nilubol D. The first detection of Senecavirus A in pigs in Thailand, 2016. Transbound Emerg Dis. 2018;65(1):285-288. https://doi.org/10.1111/tbed.12654
15. Arzt J, Bertram MR, Vu LT, Pauszek SJ, Hartwig EJ, Smoliga GR, Palinski R, Stenfeldt C, Fish IH, Hoang BH, Phuong NT, Hung VV, Vu PP, Dung NK, Dong PV, Tien NN, Dung DH. First detection and genome sequence of Senecavirus A in Vietnam. Microbiol Resour Announc. 2019;8(3):e01247-18. https://doi.org/10.1128/MRA.01247-18
16. Bennett B, Urzúa-Encina C, Pardo-Roa C, Ariyama N, Lecocq C, Rivera C, Badía C, Suárez P, Agredo M, Aguayo C, Ávila C, Araya H, Pérez P, Berrios F, Agüero B, Mendieta V, Pituco EM, de Almeida IG, Medina R, Brito B, Johow M, Ramirez VN. First report and genetic characterization of Seneca Valley virus (SVV) in Chile. Transbound Emerg Dis. 2022;69:e3462-e3468. https://doi.org/10.1111/tbed.14747
*17. SEGOB. Secretaría de Gobernación. Diario oficial de la nación. Acuerdomediante el cual se dan a conocer en los Estados Unidos Mexicanos laenfermedades y plagas exóticas y endémicas de notificación obligatoria de losanimales terrestres y acuáticos [Agreement through which mandatory notification of exotic and endemic diseases and pests of terrestrial and aquatic animals are made known in the United Mexican States]. Published November 29, 2018. Accessed December 22, 2022. https://dof.gob.mx/nota_detalle.php?codigo=5545304&fecha=29/11/ 2018#gsc.tab=0
18. Leme RA, Alfieri AF, Alfieri AA. Update on Senecavirus infection in pigs. Viruses. 2017;9(7):170. https://doi.org/10.3390/v9070170
19. Joshi LR, Mohr KA, Gava D, Kutish G, Buysse AS, Vannucci FA, Piñeyro PE, Crossley BM, Schiltz JJ, Jenkins-Moore M, Koster L, Tell R, Schaefer R, Marthaler D, Diel DG. Genetic diversity and evolution of the emerging picornavirus Senecavirus A. J Gen Virol. 2020;101(2):175-187. https://doi.org/10.1099/jgv.0.001360
20. Liu F, Wang Q, Huang Y, Wang N, Shan H. A 5-year review of Senecavirus A in China since its emergence in 2015. Front Vet Sci. 2020;7:567792. https://doi.org/10.3389/fvets.2020.567792
21. Segalés J, Barcellos D, Alfieri A, Burrough E, Marthaler D. Senecavirus A. Vet Pathol. 2017;54(1):11-21. https://doi.org/10.1177/0300985816653990
22. Hause B, Myers O, Duff J, Hesse RA. Senecavirus A in pigs, United States, 2015. Emerg Infect Dis. 2016;22(7):1323-1325. https://doi.org/10.3201/eid2207.151591
23. Maggioli MF, Fernandes MHV, Joshi LR, Sharma B, Tweet MM, Noll JCG, Bauermann FV, Diel DG. Persistent infection and transmission of Senecavirus A from carrier Sows to contact piglets. J Virol. 2019;93(21):e00819-19. https://doi.org/10.1128/JVI.00819-19
24. Turner JH, Paim WP, Maggioli MF, Peter CM, Miknis R, Talley J, Bauermann FV. Prolonged viability of Senecavirus A in exposed house flies (Musca domestica). Viruses. 2022; 14(1):127. https://doi.org/10.3390/v14010127
* Non-refereed references.
 PDF version
PDF version RIS
citation
RIS
citation