Mathew AG. 2001;3:125-129 comparison of antibiotic resistance in
Original Research (Peer-Reviewed)
A comparison of antibiotic resistance in bacteria isolated from swine herds in which antibiotics were used or excluded
Alan G. Mathew, MS, PhD; Melissa A. Beckmann, MS; Arnold M. Saxton, MS, PhD
AGM: Department of Animal Science, Brehm Bldg, 2505 River Drive, The University of Tennessee, Knoxville TN, 37996. (865) 974-7291. FAX: (865) 974-7297. Email amathew@utk.edu; MAB, AMS: Department of Animal Science, Agriculture Experiment Station, The University of Tennessee, Knoxville, Tennessee, 37996
Mathew AG, Beckmann MA, Saxton AM. A comparison of antibiotic resistance in bacteria isolated from swine herds in which antibiotics were used or excluded. J Swine Health Prod. 2001;9(3):125-129. Also available as a PDF.
Please also see errata.
Summary
Objective: To determine the effects on antibiotic resistance patterns of selected bacteria when antibiotics are used or excluded in swine.
Methods: Four herds which had been subjected to antibiotic use (AU) and three herds that had not been subjected to antibiotics (AF) were selected. From each herd, six pigs from each of four weight groups (4.5, 23, 45, and 109 kg) and five sows were randomly selected for collection of fecal samples. Non-hemolytic Escherichia coli and potential foodborne pathogens, including Salmonella spp. and E coli O157:H7, were isolated from fecal samples and tested for susceptibility to ampicillin, ceftiofur, gentamicin, oxytetracycline, and sulfamethazine, using a standardized minimum inhibitory concentration (MIC) analysis.
Results: Susceptibility patterns were different between herd types for E coli, and to a lesser extent for salmonellae. In general, Ecoli isolates from AF herds demonstrated lower MICs for ampicillin, gentamicin, oxytetracycline, and sulfamethazine. The number of resistant isolates was greater from AU herds compared to AF herds. Herd type differences were more evident for isolates from younger pigs for ampicillin, gentamicin, ceftiofur, and sulfamethazine, whereas differences were more pronounced in older pigs and sows for oxytetracycline. For salmonella, MICs for oxytetracycline and ceftiofur were greater for AU herds compared to AF herds.
Implications: Exclusion of antibiotics in swine production decreases, but does not eliminate, antibiotic resistance in E coli. Antibiotic use in swine appears to have a greater effect on resistance patterns of Ecoli than of salmonellae.
Keywords: swine, antibiotics, drug resistance, Escherichia coli, Salmonella
Received: September 12, 2000
Accepted: February 15, 2001
Antibiotics are used in livestock production to combat disease and improve animal performance. Feed-based antibiotics consistently benefit production, increasing the ability of farms to maintain profitable margins,1, 2 reducing effects of animal wastes on the environment,3 and diminishing pathogen carriage.4 However, the evolution and transfer of antibiotic resistance elements in bacteria has caused some groups to recommend restricting or banning agricultural use of antibiotics.5, 6 There is little information available regarding the impact of such restrictions on prevalence of resistant isolates. Evidence indicates that use of antibiotics in livestock production increases prevalence of resistant bacteria;7, 8, 9 however, few studies have characterized on-farm prevalence of resistant organisms in the absence of antibiotic use. Such information may be valuable for determining the potential effectiveness of restricted antibiotic use in limiting resistant bacteria in modern livestock environments. This study was designed to compare the prevalence of antibiotic resistance in bacteria associated with swine, including non-pathogenic Escherichia coli and potentialfoodborne pathogens, on farms where antibiotics were used or excluded.
Materials and methods
Seven herds from various regions of the US were selected for this study. Producers were interviewed to determine the history of antimicrobial use in each herd. Three herds, located in Iowa, New Jersey, and Kentucky, produced antibiotic-free (AF) pigs and had no history of antibiotic use for a minimum of 4, 5, and 28 years, respectively. In all AF herds, animals requiring antibiotic treatment were removed from the production site for treatment, and not returned. Four herds, two in Tennessee and two in Indiana, used antibiotics (AU) in subtherapeutic and (or) therapeutic doses. Table 1 provides information on antimicrobialuse for AU herds during the 12 months previous to the study.

In each herd, six pigs from each of four weight groups (4.5, 23, 45, and 109 kg) and five sows were randomly selected for collection of fecal samples via rectal swab. Samples were maintained in Cary-Blair medium10 on ice until arrival at the laboratory within 48 hours of collection. Samples were cultured and analyzed using standard methods for identification of E coli, E coli O157:H7, and salmonellae.
For E coli, samples were streaked onto lactose MacConkey agar and incubated overnight (37 degrees C). Pink colonies with characteristic E coli morphology were confirmed to the species by API20E biochemical analysis (Vitek bioMerieux, Syosset, New York). Isolates were further characterized on 5% sheep’s blood agar for selection of non-hemolytic strains. To detect O157:H7 isolates, samples were first incubated on sorbitol MacConkey agar (24 h, 37 degrees C), and sorbitol-negative isolates were subcultured on MacConkey MUG (4-methyllumbelliferyl-B-D-glucoronide) agar (24 h, 37 degrees C). Suspect colonies were then subjected to API20E biochemical tests, O157 antisera latex agglutination tests (DIFCO, Detroit, Michigan), and oxoid O157 antisera agglutination tests (Oxoid DR620, Ogdensburg, NewYork) for final confirmation. For isolation of salmonellae, swab tips were first vortexed in 5 mL of phosphate buffered saline (pH 7.2), and 1 mL of this mixture was cultured overnight (37 degrees C) in 5 mL of brain heart infusion (DIFCO), and subcultured first in lactose broth pre-enrichment medium for 24 h (35 degrees C), then in tetrathionate broth for 24 h (42 degrees C). Finally, the enrichment culture was streaked onto XLT4, brilliant green, and bismuth sulfite agars (DIFCO) for final confirmation. Salmonellae were subjected to serological identification using Bacto Salmonella O, H, and Vi antisera (DIFCO) to determine serovars.
For each fecal sample, a maximum of ten isolates from each bacterial group (non-pathogenic E coli, O157 E coli, and salmonella), as available, were subjected to further analysis. Susceptibility to ampicillin, ceftiofur, gentamicin, oxytetracycline, and sulfamethazine was determined using a standardized minimum inhibitory concentration (MIC) broth dilution method as outlined by the National Committee for Clinical Laboratory Standards.11 Susceptible control strains used to monitor the efficacy of each MIC test included E coli American Type Culture Collection #25922 and the swine-derived Salmonella serovar Typhimurium isolate # 798-4232 (National Animal Disease Center, USDA). Resistancewas determined at the following breakpoints: ampicillin, 32 [micro]g/mL; ceftiofur, 8 [micro]g/mL; gentamicin and oxytetracycline, 16 [micro]g/mL; and sulfameth-azine, 512 [micro]g/mL. Breakpoints for ampicillin, gentamicin, oxytetracycline, and sulphamethazine were based on human-derived breakpoints adapted for use in veterinary medicine, whereas the breakpoint for ceftiofur was based on those established for respiratory pathogens in swine.11
Statistical analysis
Susceptibility to antibiotics and the number of resistant isolates for each herd type were compared using two-way analysis of variance,12 and LSD mean separation13 was used to compare MICs and the percentage of resistant E coli between herd types and pig size groups. The individual herd served as the experimental unit to determinemain effects of antibiotic use or exclusion. All data were checked for distributional requirements prior to final analysis. Because only one AF and two AU herds were represented by the salmonella data, an ANOVA analysis was not possible. Instead, a contingency table analysis was conducted to determine differences among those herds.13
Results
Similar numbers of E coli were readily isolatedfrom all samples from both herd types. While a number of sorbitol-negative E coli were found, only 11 were confirmed to be O157 positive, representing two AU herds that were both close to cattle farms. None of the isolates demonstrated resistance to the test antibiotics. Because of the small number of isolates, and lack of isolates from AF herds, a statistical analysis was not possible for E coli O157. One hundred and forty-three salmonellae isolates were recovered from three herds (two AU herds and one AF herd ). Most of these isolates (92%) were Salmonella serovar Typhimurium, O-antigen Type B.
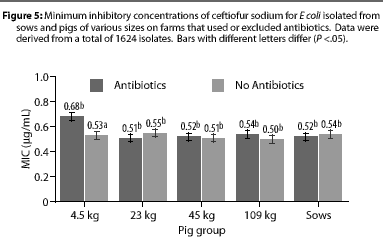
Overall, E coli from AF herds demonstrated lower MICs (P <.001) for ampicillin (Figure 1), gentamicin (Figure 2), oxytetracycline (Figure 3), and sulfamethazine (Figure 4), compared to AU herds. Resistance to ceftiofur tended to be low for all isolates, and even though differences occurred in MICs between the two herd types in isolates from the 4.5 kg pigs, all isolates were still within the range ( 2 ug per mL) considered to be susceptible to that drug (Figure 5). The differences in Ecoli susceptibility between herd types were most evident for gentamicin and ceftiofur in isolates from younger pigs. Differences between herd types were noted among almost all pig groups for ampicillin, oxytetracycline, and sulfamethazine. Fewer E coli isolates resistant to ampicillin, gentamicin, oxytetracycline, and sulfamethazine were cultured from AF herds (Figure 6). Additionally, fewer E coli isolates from AF herds demonstrated multiple resistance to the test antibiotics (P<.05), compared to AF herds (data not shown).
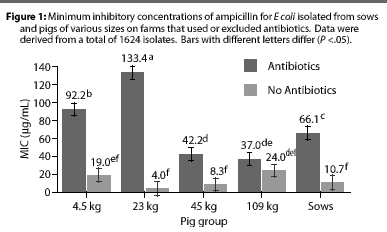
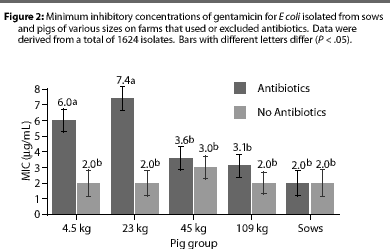
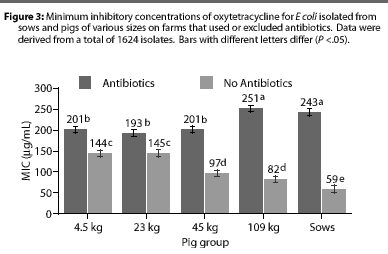
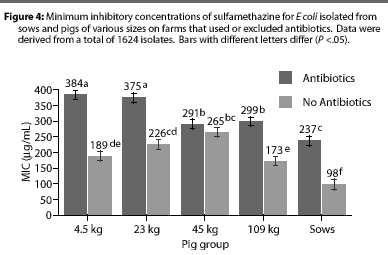

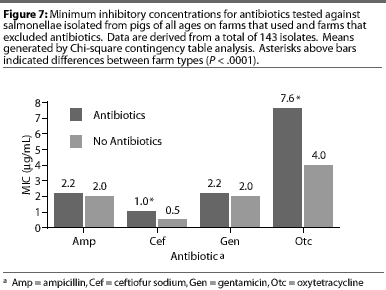
Although all Salmonella isolates were susceptible to ceftiofur (MICs <2 [micro]g per mL), the MIC was lower (Chi-square, P<.001) in the AF herd, compared to the two AU herds (Figure 7). Susceptibility of salmonellae to gentamicin was also high in all herds, although there was a trend towards higher MICs in some isolates from one AU herd (Chi-square, P =.13). Similarly, the highest MICs for oxytetracycline (Chi-square, P <.001) were found in salmonellae from AU herds. Approximately 2% of salmonellae from AU farms were determined to be resistant to oxytetracycline, on the basis of human-derived breakpoints (MIC 16 [micro]g per mL), whereas no resistant isolates were cultured from the AF herd. Susceptibility of isolates to sulfamethazine was not different (Chi-square, P =.40) between herd types, with average MICs ranging between256 and 512 [micro]g per mL for both AF and AU herds.
Discussion
While the data from this investigation could be reported several ways, we have chosen to summarize the findings by reportingmean MICs for isolates from the various sources. In evaluating the data, we determined that mean MIC was an indication of both susceptibility and prevalence of resistant organisms. In this study, resistance was not an all-or-none phenomenon occurring at only a few finite levels, but rather it occurred over a wide range of values in animal isolates. Thus, we contend that mean MIC is an appropriate measure of overall resistance of bacterial populations, and can be used to gauge effects of various production or treatment factors.
Because the criteria for resistance were based on human-derived breakpoints (all test antibiotics except ceftiofur) or on breakpoints established for respiratory pathogens (ceftiofur), it should be noted that these data may not be indicative of the clinical efficacy of all or any of the antibiotics against swine isolates.
Differences between AU and AF herds in susceptibility of E coli isolates were dependent on the antibiotic and age of pig. For example, susceptibility to oxytetracycline and ampicillin differed greatly between AU and AF herds and between pigs of varying ages, whereas susceptibility to ceftiofur differedlittle between herd types and ages of pig. In agreement with earlier studies,14,15 resistance to some antibiotics was greater in isolates from young pigs compared to isolates from older pigs and sows. The greater MICs noted for ampicillin in Ecoli from younger animals were primarily due to larger numbers of resistant organisms with breakpoints 128 [micro]g per mL. For gentamicin, greater numbers of isolates from younger pigs with breakpoints >0.5 [micro]g per mL were noted, particularly from one AU farm. Interestingly, however, a similar trend was noted for both AU and AF herds, suggesting that the greater resistance in isolates from younger animals is not necessarily associated with antibiotic use during that growth phase. It is likely, then, that genetic resistance elements are widespread among enteric bacteria, even in the absence of antibiotic use. Additionally, naturally occurring antibiotics, known to be produced by a variety of organisms,16 may provide a low level of selective pressure for maintenance of resistance elements in the farm environment. Still, MICs were greater for isolates from AU herds for all antibiotics except ceftiofur, indicating that on-farm antibiotic use likely plays a role in the susceptibility or resistance of isolates.
It should be noted that our data on Salmonella isolates
may be biased because of the necessary enrichment procedure for
recovery, which allows a few individual organisms to produce clones
with highly similar characteristics, skewing the results of the
tested population. Analysis of the salmonella data, taking into
account the number of separate positive samples, serovar differences,
and differing resistance patterns of the Salmonella isolates,
which we assumed were individual isolates, showed that our data
could represent as few as nine isolates for AF herds and 35 for
AU herds, or as many as 36 isolates for AF herds and 107 for AU
herds. While the tested pool of salmonellae is small compared
to the E coli pool, the Chi Square analysis still appears
to indicate that salmonellae may be less affected by farm use
of antibiotics, as differencesbetween herd types were noted only
for ceftiofur and tetracycline, and few isolates were resistant.
This observation is reinforced by recent challenge studies in
which we noted a marked difference in acquisitionof resistance
between
E coli and Salmonella isolates. In those studies,
resident E coli showed a much greater ability to gain resistance
under antibiotic use, compared to a Salmonella Typhimurium
challenge organism.17
In this study, excluding antibiotics from swine herds reduced the number of resistant bacteria cultured from the animals, but resistant isolates did occur in enteric bacteria, especially in young animals. Thus, imposing greater restrictions on antibiotic use in animal agriculture is likely to reduce, but not eliminate, the occurrence of resistant isolates in livestock.
Implications
- Increased restrictions on antibiotic use and (or) movement to non-antibiotic production of swine may reduce but will likely not eliminate antibiotic resistance elements (R-factors) in fecal bacteria.
- In swine, resistance patterns of E coli appear to be affected to a greater degree by antibiotic use than resistance patterns of salmonellae; thus, E coli may not be suitable sentinel organisms to indicate effects of antibiotics on resistance of the foodborne pathogen Salmonella serovar Typhimurium.
- The greater antibiotic resistance observed in bacterial isolates from young pigs is not solely the result of more antibiotic use at this stage, as similar trends were noted for farms that did not use antibiotics.
Acknowledgement
This work was funded in part by the National Pork Producer’s Council, on behalf of the National Pork Board.
References
1. Cromwell GL, Davis GW, Morrow WE, Primo RA, Borzeboom DW, Sims MD, Staisiewske EP, Ho CH. Efficacy of the antimicrobial compound U-82,127 as a growth promoter for growing-finishing pigs. J Anim Sci. 1996;74:1284-1287.
2. Stahly TS, Cromwell GL, Monegue HJ. Effects of dietary inclusion of copper and (or) antibiotics on the performance of weanling pigs. J Anim Sci. 1980;51:1347-1351
3. Roth FX, Kirchgessner M. Influence of avilamycin and tylosin on retention and excretion of nitrogen in finishing pigs. J Anim Physiol Anim Nutr. 1993;69:245-250.
4. Kyriakis SC, Sarris K, Kritas SK, Tsinas AC, Giannakopoulos C. Effect of salinomycin in the control of Clostridium perfringens type C infections in suckling pigs. Vet Rec. 1996;138:281-283.
5. World Health Organization. The medical impact of the use of antimicrobials in food animals: a report of a WHO meeting. Berlin, Germany: 1997. Document No. WHO/EMC/ZOO/97.4.
6. Brander GC. EU ban on four antibiotic feed additives. Vet Rec. 1999;144(4):104.
7. Berghash SR, Davidson JN, Armstrong JC, Dunny GM. Effects of antibiotic treatment of nonlactating dairy cows on antibiotic resistance patterns of bovine mastitis pathogens. Antimicrob Agents Chemother 1983;24(5):771-776.
8.. National Research Council Institute of Medicine. The Use of Drugs in Food Animals: Benefits and Risks. Washington, DC: National Academy Press;1998:7-10.
9. Mathew AG, Saxton AM, Upchurch WG, Chattin SE. Multiple resistance patterns of Escherichia coli isolated from high intensity swine farms. J Appl Env Microbiol. 1999;65:2770-2772.
10. Atlas RM, Snyder JW. Handbook of media for clinical microbiology. Boca Raton, Florida: CRC Press;1995:61.
11. NCCLS. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Tentative Standard. M31-T, NCCLS Vol 17(11). Villanova, Pennsylvania: National Committee for Clinical Laboratory Standards;1997:1-63.
12. SAS. SAS/STAT Users Guide (Version 6). Cary, North Carolina: SAS Institute Incorporated; 1990:23-25.
13. Steel RGD, Torrie JH. Principles and Procedures of Statistics: A Biometric Approach. 2nd ed. New York, New York: McGraw-Hill Publishing Co;1980:407-519.
14. Mathew AG, Upchurch GW, Chattin SE. Incidence of antibiotic resistance in fecal Escherichia coli isolated from commercial swine farms. J Anim Sci. 1998;76:429-434.
15. Langlois BE, Dawson KA, Cromwell GL, Stahly TS. Antibiotic resistance in pigs following a 13-year ban. J Anim Sci. 1986;62:18-32.
16. Wiener P. Experimental studies on the ecological role of antibiotic production in bacteria. Evol Ecol. 1996;10:405-421.
17. Jackson FR, Beckmann M, Mathew AG. Effect of drug combinations and regimens on antibiotic resistance in bacteria from
swine. J Anim Sci. 2000;78(suppl 1):105.