The Pestivirus genus is comprised of 4 major viral species named bovine viral diarrhea virus (BVDV) type 1, BVDV type 2, classical swine fever virus (CSFV), and border disease virus (BDV), currently reclassified as Pestivirus A, Pestivirus B, Pestivirus C, and Pestivirus D, respectively. Together with an increasing number of additional Pestivirus species detected in domestic and wild animals, at least 11 viral species are recognized within the genus and named A through K.1 Bovine viral diarrhea virus and BDV can infect multiple domestic and free-ranging wildlife species. In contrast, CSFV is restricted to members of the Suidae family.2,3 The capability of pestiviruses to cross species barriers, a high viral mutation rate, and the potential to generate persistently infected (PI) animals allow it to persist in affected animal populations. However, diverse clinical presentations may result depending on the individual immune response or from differences in the cross-protective immune response.4 While BDV is considered an infectious agent for sheep and goat disease, it can cross-infect cattle, pigs, and nondomesticated species.5,6 Border disease is a viral disease associated with reproductive manifestations including abortions, fetal mummifications, stillbirths, barren ewes, birth of weak and PI lambs, abnormal body conformation, and immunosuppression. The seroprevalence rates in sheep vary depending on geographic regions and animal husbandry.7 Morbidity and mortality rates vary with age or stage of infection, strain virulence, and the infected host species.8 Transmission of BDV to pigs is possible and most likely occurs through contact with PI animals, albeit the source of viral infection cannot always be determined.9 Studies in the Netherlands described BDV as the predominant pestivirus infecting pig populations where BDV was isolated.10,11 Clinical presentations are usually mild; nonetheless, they may range from asymptomatic to clinically severe. Moreover, congenital transmission in piglets and hemorrhagic lesions in pigs have also been previously reported.10,12 This case report details the detection and characterization of BDV infection in piglets with severe clinical signs.
Animal care and use
This study was conducted at the Mexico-United States Commission for Prevention of Foot-and-Mouth Disease and Other Exotic Animal Diseases (CPA) according to good production practices in pig farms manual implemented by the Ministry of Agriculture and Rural Development.
Case description
The affected farm was in Tlaxcala, Mexico. The rural farm kept a total of 139 Pietrain × Yorkshire crossbred pigs under a semi-intensive production farming system, where the breeding herd was kept outside, allowing them to feed on natural vegetation in fenced enclosures, and piglets were housed in indoor pens. Diagnosis of infectious pathogens and vaccination protocols were poorly performed; therefore, the epidemiological status of endemic diseases was unknown. Over 6 days in August 2021, fifteen 45-day-old piglets developed clinical signs including fever, anorexia, cachexia, cyanosis, prostration, and death. Previous close contact with ruminants is unknown, and no other small ruminants or cattle were housed on the farm. Upon the onset of clinical signs, sick animals were isolated in separate pens off-site. Other biosecurity strategies were implemented including cleaning and disinfection of all areas, control protocols for entry and exit, and use of personal protective clothing.
Necropsy findings from 2 animals included hemorrhagic lung lesions and fibrosis, pleuritis, and petechial hemorrhages in the jejunum, ileum, bladder, and kidney surface epithelium. Four serum samples from sick animals and tissue samples of brain, tonsil, kidney, spleen, and mesenteric ganglia from 1 dead animal were collected and submitted for diagnosis to the Immunology, Cellular and Molecular Biology Laboratory at CPA and reported as case number CPA-12362-21. Prior to the disease event, no evidence of related clinical manifestations was registered on the farm or the neighboring farms.
Diagnosis and laboratory findings
Initially, the differential diagnosis included CSFV, African swine fever virus (ASFV), and pseudorabies virus (PRV), which were ruled out by negative real-time quantitative reverse transcriptase- polymerase chain reaction (qRT-PCR) results. Subsequently, qRT-PCR was performed to detect additional viruses that display similar clinical signs, such as porcine epidemic diarrhea virus (PEDV), porcine circovirus type 2 (PCV-2), porcine circovirus type 3 (PCV-3), transmissible gastroenteritis virus (TGEV), and porcine reproductive and respiratory syndrome virus (PRRSV); results were negative. End-point RT-PCR was used to assess BDV and BVDV presence in tissue samples, and BDV-positive results were obtained from the spleen, kidney, tonsil, and mesenteric ganglia tissue samples.
In addition, a pool of tissue samples was submitted to the National Center for Diagnostic Services in Animal Health (CENASA) for complementary qRT-PCR and polymerase chain reaction (PCR) testing for porcine parvovirus (PPV), Senecavirus A (SVA), porcine rubula- virus (PoRV), influenza A virus (IAV), Actinobacillus pleuropneumoniae, Brachyspira hampsonii, Brachyspira hyodysenteriae, Erisypelothrix rhusiopathiae, Mycoplasma, Mycoplasma hyopneumoniae, Pasteurella multocida, and Salmonella. All tests were negative except for PPV.
Virus isolation attempts from BDV- positive tissue samples, performed under the procedure described in the Manual of Diagnostic Tests and Vaccines for Terrestrial Animals13 at the Biosafety Level 3 Cell Culture Laboratory at CPA were unsuccessful. Serum from animals positive for BDV by RT-PCR was further analyzed using a virus neutralization test (VNT) and enzyme-linked immunosorbent assay (ELISA) for the presence of specific antibodies; negative results were obtained from both assays.
Due to BDV-positive pigs and new mortality cases on the described farm, and in accordance with epidemiological surveillance, an examination was carried out on the farm 3 days after BDV was first detected. Five whole-blood samples from clinically healthy animals and tonsil, liver, kidney, spleen, and mesenteric ganglia samples from 1 dead piglet were collected and submitted to the CPA for viral testing, with results reported in case number CPA-12574-21. The piglet that presented with clinical disease and death, similar to those from the initial report, was immediately diagnosed as BDV positive using end-point RT-PCR. Border disease virus was detected in 3 of 5 whole-blood samples using end-point RT-PCR. Further, BDV RNA was detected in mesenteric ganglia and liver samples. However, attempted BDV isolation from the collected tissue samples was unsuccessful. Serological testing for BDV-specific antibodies by ELISA and VNT was negative. Similarly, samples were negative for CSFV, ASFV, PRV, PEDV, PCV-2, PCV-3, TGEV, and PRRSV using the qRT-PCR technique. Subsequently, a pool of tissue samples was submitted for diagnosis of PPV, SVA, PoRV, IAV, A pleuropneumoniae, B hampsonii, B hyodysenteriae, E rhusiopathiae, Mycoplasma, M hyopneumoniae, P multocida, and Salmonella. The pool of tissue samples was positive for PPV and Mycoplasma using PCR.
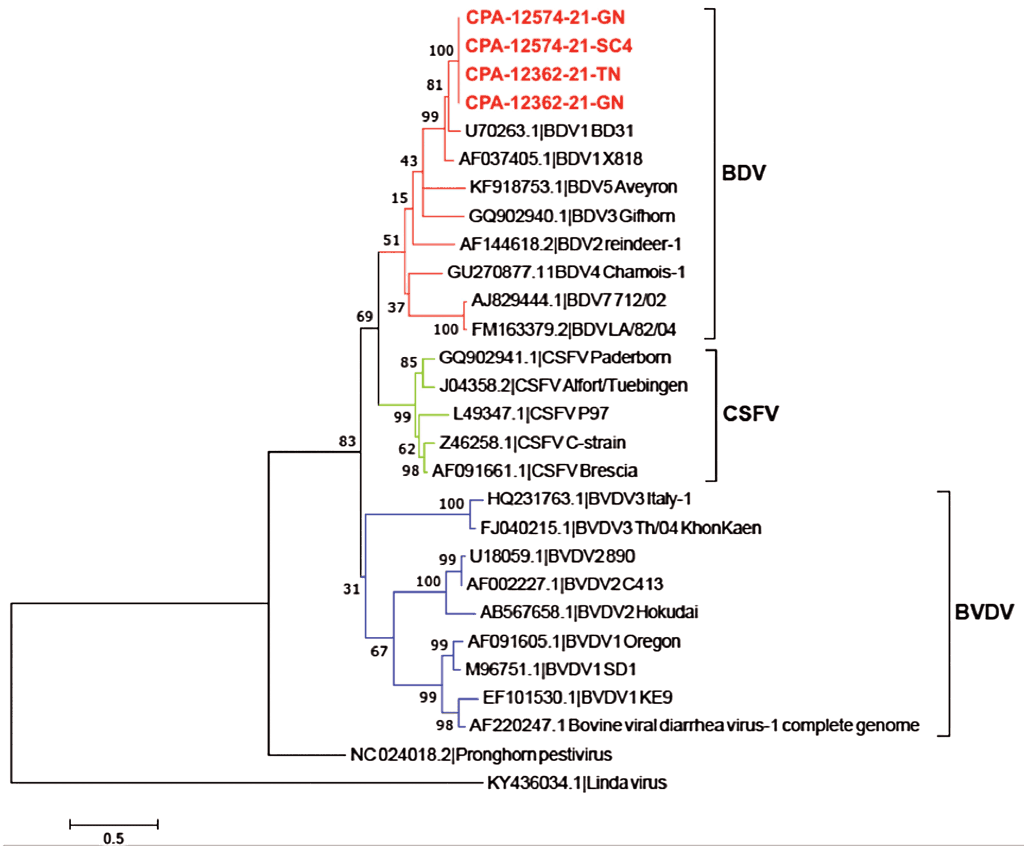
For further characterization of BDV from these cases, mesenteric ganglia and tonsil samples from case CPA-12362-21 and mesenteric ganglia and whole-blood samples belonging to case CPA-12574-21 were selected for additional analysis. Positive RT-PCR products from each case were sequenced by the Sanger method. The 4 partial Npro nucleotide sequences were individually deposited in GenBank under accession numbers OK667067, OK667068, OK667069, and OK667070. Subsequent phylogenetic analysis indicated that evaluated sequences were clustered within the BDV-1 genotype (Figure 1).
Discussion
Border disease virus is reported globally as an important pathogen with at least 8 genotypes, from BDV-1 to BDV-8.14 Detection in diverse species of even-toed ungulates, including sheep, goats, cattle, chamois, and pigs, has been previously reported.15-17 Border disease virus infection in sheep produces clinical signs ranging from mild to severe including reproductive failure, congenital disorders, and abnormal body conformation.18 In addition, congenital infection occurring during the first half of gestation may lead to abortion and stillbirth, the birth of lambs with malformations, and the birth of PI animals if BDV infection occurs before day 60 of gestation.19 These animals represent the main source of infection and maintenance of BDV in the animal population.20 Fetal death may occur at any stage of gestation. However, it is more common in fetuses infected early in gestation.15 Severity of clinical signs depend on the timing of infection during pregnancy, the virulence of the infecting strain, and the susceptibility of the species infected.15 Seroprevalence may vary from 5% to 90% among sheep populations depending on the region surveyed.17,21
Mexico has been recognized as CSFV free since 2015; however, active epidemiological surveillance is maintained to detect any indication of CSFV infection.22,23 Therefore, serological assays have been conducted to determine the prevalence of pestivirus infections in pigs. The prevalence of BDV antibodies was investigated in pigs nationwide from 2011 to October 2021 revealing an estimated 41.17% seroprevalence.24 Likewise, during a national screening for pestivirus in cattle, 3 cases were found to be BDV positive; genetic characterization typed the Mexican strains as BDV-1.5 These findings highly suggest BDV circulation in pig and cattle populations in Mexico, probably due to natural infection through close contact among ruminants and pigs since it appears to be the most crucial risk factor for interspecies transmission.25 Conversely, no BDV seroconversion was detected in this study. This is due to serum sample collection occurring in an early stage of the BDV infection; therefore, no detectable antibodies were produced by the time of sampling. Previous studies of experimentally BDV-inoculated sows showed seroconversion after 3 weeks post inoculation.26
Natural and experimental infection studies have demonstrated the susceptibility of domestic pigs to BDV strains. Border disease virus infection in pigs leading to mild or inapparent manifestations has been described elsewhere.9,26 One study showed that BDV-infected pigs with no clinical signs and no histopathological lesions could shed the virus through oronasal secretions from 3 to 7 days post infection and became viremic at 3 to 14 days post infection.17 In 1996, the Frijters strain was isolated from congenitally infected piglets and genetically characterized as a BDV strain able to infect pigs and is circulating among large populations in Europe.10 Roehe et al12 detected a virus genetically more related to BDV than CSFV or BVDV from a severe clinical manifestation in weaned pigs showing hemorrhagic lesions at necropsy. Nonetheless, an association among histopathological lesions and the presence of viral antigen is required to confirm the causative agent. Similarly, in northern and western France, the use of a BDV-contaminated vaccine elicited eyelid edema, locomotor disorders, decay, and spontaneous death in piglets and sows; at necropsy, hemorrhagic lesions were similar to those observed with CSFV. In addition, these animals showed persistent infection and immunotolerance.27
Our study describes the detection of BDV in mesenteric ganglia, tonsil, and blood samples from pigs with severe clinical disease suggesting the BDV infection was present in the surveyed animals. We performed sequencing and genetic characterization by phylogenetic inference using Npro sequence in all RT-PCR detected BDV strains, which revealed a close relationship to the BD31 strain (Figure 1). This was similar to the characterized BDV strain detected on a pig farm with no ruminants in Japan.28 The BDV-1 genotype has also been detected as the circulating BDV strain in the United States, the United Kingdom, Australia, and New Zealand.29-31
Border disease virus has been detected in serum samples from cattle in Mexico.5 Serological evidence of BDV infection in pigs has also been recorded.24 No virus isolation was obtained in this study, which is similar to other studies among the surveyed populations.28,32
At the same time, PPV was detected on both sets of tissues tested. Porcine parvovirus is considered endemic in swine populations worldwide and one of the major viral pathogens causing reproductive failure.33 Despite its detection, PPV is mainly associated with reproductive disorders summarized under the acronym SMEDI (stillbirth, mummifications, embryonic death, and infertility), with clinical disease restricted to pregnant sows or gilts. In piglets, PPV infection does not cause clinical disease.34 Moreover, the immunosuppression caused by BDV and PPV can increase the risk of opportunistic infections.15 The detection of ubiquitous Mycoplasma species in surveyed samples is not unexpected; however, it might be associated with an immunosuppressive event.35
In this case, the clinical disease presentation cannot only be associated with BDV infection since the lack of serological assay evidence and absence of pathological evaluations prevented us from determining BDV as the causative agent post event. Nevertheless, opportunistic pathogens could be involved in the severe clinical disease and should be considered.
Finding BDV in the national swine population has relevant implications in a country where CSFV eradication has been achieved as serological tests will not differentiate among BDV, BVDV, or CSFV infections.28 This is the first report of BDV in pigs in Mexico, and BDV-positive serology reinforces the suggestion that BDV can be considered an endemic virus. The latter highlights the need for implementation of accurate swine diagnostic tests able to detect and discriminate among pestiviruses and other pathogens with similar pathologies to determine the definitive cause of disease. Furthermore, surveys are needed to determine the occurrence of BDV in pigs and the impact on swine health and production.
Implications
- BDV was detected in seronegative pigs from Mexico.
- For seronegative domestic pigs, BDV remains a potential risk.
- Detecting BDV transmission in domestic pigs can be diagnostic challenge.
Acknowledgments
Conflict of interest
None reported.
Disclaimer
Scientific manuscripts published in the Journal of Swine Health and Production are peer reviewed. However, information on medications, feed, and management techniques may be specific to the research or commercial situation presented in the manuscript. It is the responsibility of the reader to use information responsibly and in accordance with the rules and regulations governing research or the practice of veterinary medicine in their country or region.
References
1. King AMQ, Lefkowitz EJ, Mushegian AR, Adams MJ, Dutilh BE, Gorbalenya AE, Harrach B, Harrison RL, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert ML, Rubino L, Sabanadzovic S, Sanfaçon H, Siddell SG, Simmonds P, Varsani A, Zerbini FM, Davison AJ. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2018). Arch Virol. 2018;163(9):2601-2631. https://doi.org/10.1007/s00705-018-3847-1
2. Becher P, Orlich M, Shannon AD, Horner G, König M, Thiel HJ. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J Gen Virol. 1997;78(6):1357-1366. https://doi.org/10.1099/0022-1317-78-6-1357
3. Blome S, Staubach C, Henke J, Carlson J, Beer M. Classical swine fever – an updated review. Viruses. 2017;9(4):86. https://doi.org/10.3390/v9040086
4. Becher P, Avalos Ramirez R, Orlich M, Cedillo Rosales S, König M, Schweizer M, Stalder H, Schirrmeier H, Thiel HJ. Genetic and antigenic characterization of novel pestivirus genotypes: Implications for classification. Virology. 2003;311(1):96-104. https://doi.org/10.1016/s0042-6822(03)00192-2
5. Gómez-Romero N, Basurto-Alcántara FJ, Verdugo-Rodríguez A, Lagunes-Quintanilla R, Bauermann FV, Ridpath JF. Detection of border disease virus in Mexican cattle. Transbound Emerg Dis. 2018;65(1):267-271. https://doi.org/10.1111/tbed.12641
6. Nettelton PF, Entrican G. Ruminant pestiviruses. Br Vet J. 1995;151:615-642. https://doi.org/10.1016/s0007-1935(95)80145-6
7. Righi C, Petrini S, Pierini I, Giammarioli M, De Mia GM. Global distribution and genetic heterogeneity of border disease virus. Viruses. 2021;13(6):950. https://doi.org/10.3390/v13060950
8. Chappuis G, Brun A, Kato F, Dauvergne M, Reynaud G, Duret C. Études sérologiques et immunologiques réalisées à la suite de l’isolement d’un pestivirus dans un Foyer Ovin chez des Moutons de l’Aveyron [Serological and immunological studies carried out following the isolation of a pestivirus in a sheep outbreak in Aveyron]. In: Espinasse J, Savey M, eds. Pestivirose des Ovinset des Bovins: Nouvelles Connaissances, Utilisation pour une Stratégie de Contrôle, Journées Nationales de la Société Française de Buiatrie et de son Groupe d’Étudesur la Pathologie des Ovins et des Caprins (GEPOC). Societe Francaise de Buiatrie; 1986:55-65.
9. Rosell R, Cabezón O, Pujols J, Domingo M, Muñoz I, Núñez JI, Ganges L. Identification of a porcine pestivirus as a border disease virus from naturally infected pigs in Spain. Vet Rec. 2014;174(1):18. https://doi.org/10.1136/vr.101920
10. Vilcek S, Belák S. Genetic identification of pestivirus strain Frijters as a border disease virus from pigs. J Virol Methods. 1996;60(1):103-108. https://doi.org/10.1016/0166-0934(96)02031-9
11. de Smit AJ, Eblé PL, de Kluijver EP, Bloemraad M, Bouma A. Laboratory decision-making during the classical swine fever epidemic of 1997-1998 in The Netherlands. Prev Vet Med. 1999;42(3-4):185-199. https://doi.org/10.1016/s0167-5877(99)00075-6
12. Roehe PM, Woodward MJ, Edwards S. Characterization of p20 gene sequences from a border disease-like pestivirus isolated from pigs. Vet Microbiol. 1992;33(1-4):231-238. https://doi.org/10.1016/0378-1135(92)90051-t
*13. World Organization for Animal Health. Chapter 3.8.1. Border Disease. In: Manual of diagnostic tests and vaccines for terrestrial animals. 8th ed. OIE; 2018:5-6. Accessed November 12, 2021. https://www.oie.int/fileadmin/Home/esp/Health_standards/tahm/3.08.01_Enfermedad_frontera.pdf
14. Peletto S, Caruso C, Cerutti F, Modesto P, Zoppi S, Dondo A, Acutis PL, Masoero L. A new genotype of border disease virus with implications for molecular diagnostics. Arch Virol. 2016;161(2):471-477. https://doi.org/10.1007/s00705-015-2696-4
15. Nettleton PF, Gilray JA, Russo P, Dlissi E. Border disease of sheep and goats. Vet Res. 1998;29(3-4):327-340.
16. De Mia GM, Greiser-Wilke I, Feliziani F, Giammarioli M, De Giuseppe A. Genetic characterization of a caprine Pestivirus as the first member of a putative novel pestivirus subgroup. J Vet Med B Infect Dis Vet Public Health. 2005;52(5):206-210. https://doi.org/10.1111/j.1439-0450.2005.00850.x
17. Cabezón O, Rosell R, Velarde R, Mentaberre G, Casas-Díaz E, Lavín S, Marco I. Border disease virus shedding and detection in naturally infected Pyrenean chamois (Rupi-capra pyrenaica). J Vet Diagn Invest. 2010;22(5):744-747. https://doi.org/10.1177/104063871002200514
18. Berriatua E, Barandika JF, Aduriz G, Hurtado A, Estévez L, Atxaerandio R, García-Pérez AL. Flock-prevalence of border disease virus infection in Basque dairy-sheep estimated by bulk-tank milk analysis. Vet Microbiol. 2006;118(1-2):37-46. https://doi.org/10.1016/j.vetmic.2006.06.013
19. Nettleton PF. Pestivirus infections in ruminants other than cattle. Rev Sci Tech. 1990;9(1):131-150. https://doi.org/10.20506/rst.9.1.485
20. Dhahir HS, Talb OQ, Asim M. Preliminary study of seroprevalence of border disease virus (BDV) among sheep and goats in Mosul City, Iraq. Adv Anim Vet Sci. 2019;7(7):566-569. https://doi.org/10.17582/journal.aavs/2019/7.7.566.569
21. Kaiser V, Nebel L, Schüpbach-Regula G, Zanoni RG, Schweizer M. Influence of border disease virus (BDV) on serological surveillance within the bovine virus diarrhea (BVD) eradication program in Switzerland. BMC Vet Res. 2017;13(1):21. https://doi.org/10.1186/s12917-016-0932-0
*22. World Organization for Animal Health. Recognition of the classical swine fever status of members. Resolution No. 20. Published 2021. Accessed November 29, 2021. https://www.oie.int/app/uploads/2021/05/a-r20-2021-csf.pdf
*23. World Organization for Animal Health. Recognition of the sanitary status of member countries with respect to foot-and-mouth disease. Resolution No. 17. Published 2015. Accessed November 30, 2021. https://www.oie.int/app/uploads/2021/03/e-reso-2015-public.pdf
*24. Servicio Nacional de Salud, Inocuidad y Calidad Agroalimentaria. Sistema de información nacional de enfermedades exóticas y emergentes (SINEXE) [National information system for exotic and emergent diseases (SINEXE)]. Published 2016. Accessed October 25, 2021. https://www.gob.mx/senasica/acciones-y-programas/sistema-de-informacion-nacional-de-enfermedades-exoticas-y-emergentes-sinexe
25. Strong R, La Rocca SA, Ibata G, Sandvik T. Antigenic and genetic characterization of border disease viruses isolated from UK cattle. Vet Microbiol. 2010;141(3-4):208-215. https://doi.org/10.1016/j.vetmic.2009.09.010
26. Leforban Y, Vannier P, Cariolet R. Protection of piglets born from ruminant pestivirus experimentally infected sows, and their contacts, to the challenge with hog cholera virus. Ann Rech Vet. 1992;23(1):73-82.
27. Vannier P, Leforban Y, Carnero R, Cariolet R. Contamination of a live virus vaccine against pseudorabies (Aujeszky’s disease) by an ovine pestivirus pathogen for the pig. Ann Rech Vet. 1988;19(4):283-290.
28. Kawanishi N, Tsuduku S, Shimizu H, Ohtani Y, Kameyama K, Yamakawa M, Tsutsui T, Matsuura K, Ohashi S, Isobe T, Yamada S. First isolation of border disease virus in Japan is from a pig farm with no ruminants. Vet Microbiol. 2014;171(1-2):210-214. https://doi.org/10.1016/j.vetmic.2014.03.032
29. Sullivan DG, Chang DJ, Akkina RK. Genetic characterization of ruminant pestiviruses: Sequence analysis of viral genotypes isolated from sheep. Virus Res. 1997;47:19-29. https://doi.org/10.1016/s0168-1702(96)01402-5
30. Becher P, Shannon AD, Tautz N, Thiel HJ. Molecular characterization of border disease virus, a pestivirus from sheep. Virology. 1994;198(2):542-551. https://doi.org/10.1006/viro.1994.1065
31. Vilcek S, Björklund HV, Horner GW, Meers J, Belák S. Genetic typing of pestiviruses from New Zealand. N Z Vet J. 1998;46(1):35-37. https://doi.org/10.1080/00480169.1998.36048
32. Ridpath JF, Neill JD. Challenges in identifying and determining the impacts of infection with pestiviruses on the herd health of free-ranging cervid populations. Front Microbiol. 2016;7:921. https://doi.org/10.3389/fmicb.2016.00921
33. Mengeling WL, Lager KM, Vorwald AC. The effect of porcine parvovirus and porcine reproductive and respiratory syndrome virus on porcine reproductive performance. Anim Reprod Sci. 2000;60-61:199-210. https://doi.org/10.1016/S0378-4320(00)00135-4
34. Zeeuw EJL, Leinecker N, Herwig V, Selbitz H-J, Truyen U. Study of the virulence and cross-neutralization capability of recent porcine parvovirus field isolates and vaccine viruses in experimentally infected pregnant gilts. J Gen Virol. 2007;88(2):420-427. https://doi.org/10.1099/vir.0.82302-0
35. Thacker EL, Minion FC. Mycoplasmosis. In: Zimmerman J, Karriker L, Ramirez A, Schwartz K, Stevenson G, eds. Diseases of Swine. 10th ed. Wiley-Blackwell; 2012:779-797.
* Non-refereed references.
 PDF version
PDF version RIS
citation
RIS
citation