| Brief communication | Peer reviewed |
Cite as: Lopez WA, Angulo J, Zimmerman JJ, et al. Porcine reproductive and respiratory syndrome monitoring in breeding herds using processing fluids. J Swine Health Prod. 2018;26(3):146-150. https://doi.org/10.54846/jshap/1055
Also available as a PDF.
SummaryProcessing fluids (PF), the serosanguinous fluid recovered from piglet castration and tail docking, were used for porcine reproductive and respiratory syndrome virus (PRRSV) infection assessment. Processing fluid samples from four breed-to-wean herds were compared with standard sampling protocols, demonstrating PRRSV RNA detection in PF at greater frequency than standard schemes. | ResumenLos fluidos de procesamiento (PF por sus siglas en inglés), el fluido serosanguíneo recuperado de la castración de lechones y corte de cola, fueron utilizados para monitorear la infección del virus del síndrome reproductivo y respiratorio porcino (PRRSV). Se compararon muestras de fluidos de procesamiento de cuatro hatos de cría a destete con protocolos de muestreo estándar, demostrando la detección del RNA del PRRSV en PF a una frecuencia mayor que en los esquemas estándar. | ResuméLes fluides de traitement (FT), le fluide séro-sanguignolant récupéré lors de la castration et de la taille de la queue des porcelets, ont été utilisés pour évaluer l’infection par le virus du syndrome reproducteur et respiratoire porcin (VSRRP). Des échantillons de FT de quatre troupeaux de type reproducteur-sevrage ont été comparés avec les protocoles standards d’échantillonnage, démontrant la détection d’ARN du VSRRP dans les FT à une fréquence plus élevée que les façons standards. |
Keywords: swine, porcine reproductive and respiratory syndrome virus, monitoring, surveillance, processing fluids, PRRSV
Search the AASV web site
for pages with similar keywords.
Received: July 14, 2017
Accepted: November 3, 2017
Swine producers face ongoing challenges related to the detection and management of infectious diseases. In particular, porcine reproductive and respiratory syndrome (PRRS) is an economically significant problem, costing US producers more than 1 billion USD per year.1 A milestone in the control and elimination of PRRS virus (PRRSV) in production systems is the interruption of the transmission cycle in breeding herds and the production of PRRSV-free piglets at weaning.2 Tracking progress towards this goal can only be accomplished through routine diagnostic monitoring.
The current industry standard for monitoring PRRSV in breeding herds consists of testing serum samples monthly from 30 randomly-selected weaning-age piglets (pooled by five) for PRRSV RNA. Breeding herds are defined as “stable” after four consecutive negative monthly tests.3 This monitoring plan assumes that PRRSV cannot remain in breeding herds at a prevalence < 10% over a period of 90 days, and that the true PRRSV status of the breeding herd can be accurately inferred by testing suckling pigs. However, cases of breeding herds detecting PRRSV shortly after achieving stability have been reported.4 Likewise, near-zero PRRSV prevalence has been documented in endemically-infected breeding herds.4-7
These observations are evidence that the assumptions upon which the current monitoring plan is based are not sufficiently robust to provide reliable results. Thus, there is a clear need for improved PRRSV monitoring systems. The current monitoring scheme could be improved upon by testing higher numbers of individual piglets at a higher testing frequency. However, collecting blood samples from piglets is time consuming, requires two trained persons, and causes additional piglet stress. These practical and economic constraints render this option unsatisfactory for most commercial production systems.
Aggregate (population) samples, eg, oral fluids, are practical options for infectious disease monitoring of swine populations. Oral-fluid testing was introduced into the swine industry in 2010 and has been widely implemented in monitoring and surveillance systems.5-11 However, the collection of oral fluids from suckling piglets has not been proven to be practical. Alternatively, a largely unexplored option for PRRSV surveillance in the breeding herd and suckling piglet populations is the use of “processing fluids.” An aggregate sample easily collected by farm staff, processing fluid samples are defined as the serosanguinous fluid recovered at the time of castration and tail docking, ie, piglet processing. The purpose of this pilot study was to describe the collection of processing fluids in commercial herds and evaluate their use in PRRSV monitoring.
Materials and methods
This study was approved by the Iowa State University Institutional Animal Care and Use Committee under protocol No. 6-17-8547-S.
Study design
Twelve samplings were performed in four breed-to-wean herds at different time points within 27 weeks of their most recent clinical PRRS episode. Each sampling consisted of one aggregate processing fluid sample, composed of the fluids from all piglets processed that day, and serum samples from 30 piglets conveniently selected from the same population of processed piglets, targeting the weak and fall behind animals and including males and females. All processing fluid samples and serum samples were tested for PRRSV RNA (serum samples were tested in pools of five). Selected processing fluid samples (n = 5) were submitted for PRRSV ORF-5 sequencing and for detection of PRRSV antibody. Testing was performed at the Iowa State University Veterinary Diagnostic Laboratory (ISU VDL) using routine test methods. Two farms (matching sets 1, 2, 4, 6, and 10) were vaccinating piglets with Fostera PRRS (Zoetis, Parsippany, New Jersey) immediately after processing.
Sample collection and matching
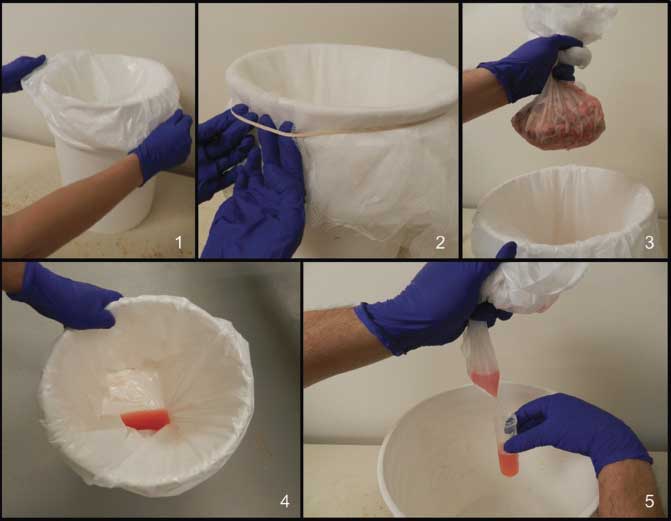
Processing fluids were collected by placing a disposable plastic bag in a clean plastic bucket and then covering the top of the bucket with disposable gauze, ie, cheese cloth. A rubber band around the mouth of the bucket held the plastic bag and gauze firmly in place, but the gauze was placed with sufficient slack so as to create a concave cavity in which to hold the tissues (Figure 1). At the end of piglet processing, the gauze and tissues were removed from the bucket, after which the processing fluids were transferred from the plastic bag to a sterile 50 mL conical centrifuge plastic tube. The number of piglets that contributed to the processing fluid sample was recorded for each sampling.
Figure 1: Steps for collecting processing fluids. Panel 1: Plastic bag in clean bucket. Panel 2: Cheese cloth placed over mouth of bucket to hold tissues and allow fluid to pass through to plastic bag. Panel 3: Tissues are removed after collection. Panel 4: Processing fluid recovered in plastic bag: Panel 5: Fluid decanted from plastic bag into tube.
Blood samples were collected from a convenience sample of 30 piglets in the same room at the time processing fluids were obtained. Blood was obtained using single-use serum separation tubes and needles and standard procedures for the restraint of piglets.
Blood samples and processing fluids were refrigerated immediately after collection and submitted for testing within 24 hours. Processing fluids were tested individually; serum samples were tested in pools of five. Thus, each “matched sampling set” consisted of one processing-fluid sample and six pooled-serum samples.
Diagnostic testing
Diagnostic testing was performed at the ISU VDL using assays routinely used for swine serum samples in the case of serum and oral fluids (high volume extraction protocol) for processing fluids. All processing fluids samples (n = 12) and pooled serum samples (n = 72) were tested for PRRSV RNA using the Applied Biosystems TaqMan kit for North American and European PRRS virus RNA detection (Thermo Fisher Scientific, Waltham, Massachusetts). In addition, five matched sampling sets (5, 7, 8, 9, 10) were conveniently selected to be tested for PRRSV antibody testing using the IDEXX PRRS X3 Ab ELISA test (IDEXX Laboratories, Westbrook, Maine), and five matched sampling sets (2, 3, 4, 5, 7) were submitted for PRRS ORF-5 sequencing (Sanger method).12
Results
Processing fluids were obtained on all attempts (Table 1), yielding a median volume of 49.0 mL (range 30.0 to 110.0 mL) of fluids. The median number of piglets that contributed to processing fluids was 256 (ranging from 174 to 650) and the average volumes of processing fluid per litter and per pig were 2.17 mL (1.22 to 2.67) and 0.186 mL (0.097 to 0.276), respectively. The age of sampled piglets ranged from 3 to 5 days.
Table 1: Volume of processing fluids retrieved from piglets at processing time (castration and tail docking) at each sampling point
| Sampling set* | Processing fluids retrieved volume (mL) | Litters in sample | Piglets in sample | Average processing fluids volume† | |
|---|---|---|---|---|---|
| Per litter (mL) | Per pig (μL) | ||||
| 1 | 30 | 21 | 262 | 1.43 | 115 |
| 2 | 45 | 21 | 250 | 2.14 | 180 |
| 3 | 48 | 18 | 174 | 2.67 | 276 |
| 4 | 50 | 20 | 226 | 2.50 | 221 |
| 5 | 55 | 25 | 265 | 2.20 | 208 |
| 6 | 45 | 37 | 466 | 1.22 | 97 |
| 7 | 80 | 35 | 438 | 2.29 | 183 |
| 8 | 110 | 50 | 650 | 2.20 | 169 |
| 9 | 90 | 37 | 481 | 2.43 | 187 |
| 10 | 45 | 17 | 221 | 2.65 | 204 |
| 11 | 32 | 17 | 177 | 1.88 | 181 |
| 12 | 50 | 21 | 233 | 2.38 | 215 |
| Totals | 680 | 319 | 3843 | 2.17 | 186 |
* Each ‘Sampling set’ consists of one processing-fluid sample and 30 serum samples (tested in six pools of five) taken from the same piglet population, on the same day of piglet processing at each sampling point.
† This table shows only the volume of processing fluid retrieved in each sampling set.
Ten of 12 processing-fluid samples (83.33%) tested positive for PRRSV RNA by real time reverse transcription polymerase chain reaction (rRT-PCR) with cycle threshold (Ct) values ranging from 22 to 35 (Table 2). Eleven of 72 (15.27%) pooled serum samples tested positive. Eight of 12 matching sets (66.66 %) had at least one positive pooled serum sample (of six pools of five samples) test positive for PRRSV PCR (Table 2).
Table 2: Qualitative result of PRRSV rRT-PCR tests in processing fluids and matching serum samples and timeline of PRRS outbreak and whole-herd exposure
| Sampling set | Time between PRRS outbreak and sampling (weeks) | Time from whole-herd exposure to MLV or FVE (weeks) | Result of PRRSV rRT-PCR | |||
|---|---|---|---|---|---|---|
| Processing fluids | Serum samples | |||||
| Ct value* | Test result | Ct value* | Test result | |||
| 1 | 6.0 | FVE: 1.0 | 31.7 | Positive | 23.0 | Positive |
| 2 | 5.4 | MLV: 5.4 | 28.4 | Positive | 20.1 | Positive |
| 3 | 7.9 | MLV: 6.9 | 30.1 | Positive | 27.0† | Positive |
| 4 | 9.6 | MLV: 9.6 | 25.6 | Positive | 23.7 | Positive |
| 5 | 11.9 | MLV: 10.9 | 22.7 | Positive | 27.6 | Positive |
| 6 | 8.0 | MLV: 2.0 | 29.2 | Positive | 25.0† | Positive |
| 7 | 20.0 | MLV: 9.0 | 34.1 | Positive | 40.0 | Negative |
| 8 | 21.1 | MLV: 10.1 | 35.2 | Positive | 40.0 | Negative |
| 9 | 22.0 | MLV: 11.0 | 26.4 | Positive | 27.9 | Positive |
| 10 | 11.4 | MLV: 5.4 | 30.2 | Positive | 31.1† | Positive |
| 11 | 16 | MLV: 15 | 40.0 | Negative | 40.0 | Negative |
| 12 | 27.1 | MLV: 26.1 | 40.0 | Negative | 40.0 | Negative |
* Samples with Ct values < 37 are considered positive samples and those with Ct values ≥ 37 are considered negative samples.
† These Ct values represent the average Ct values of two positive pools of serum samples out of the six pools tested from the correspondent sampling set.
PRRSV = Porcine reproductive respiratory syndrome virus; rRT-PCR = Real time reverse transcription polymerase chain reaction; PRRS = Porcine reproductive respiratory syndrome; MLV = modified live vaccine (PRRSV); FVE = field virus exposure; Ct = Cycle threshold from the rRT-PCR assay.
All processing fluids submitted for serology tested positive for PRRSV antibody (n = 5), with sample-to-positive (S:P) ratios of 1.50, 1.01, 2.50, 0.42, and 0.92, respectively. Likewise, it was possible to sequence the PRRSV ORF-5 from processing fluids in all attempts (n = 5). All five cases had a 100% nucleotide sequence homology when comparing processing fluids and blood samples from the same population. In all cases, the ORF-5 sequence was identified as wild-type PRRSV.
Discussion
This study described the process of collecting processing fluids from 3- to 5-day-old piglets and provided initial data on the use of processing fluids for PRRSV monitoring. Recovering processing fluids from testicles and tails of 3- to 5-day-old piglets was practical and convenient for farm staff under field conditions. We emphasize the importance of the biosecurity measures that were used (disposable materials described in Figure 1) to avoid contamination of fluids with nucleic acid present in the farm environment.
The current procedures for PRRSV ORF-5 sequencing and antibody detection in serum samples were compatible with processing fluids. This was not unexpected but required verification. Testing results indicated that the likelihood of PRRSV RNA detection in processing fluids was greater than the likelihood of detecting PRRSV RNA in 30 matched serum samples (tested in pools of five) from the same population. Thirty serum samples were used as a comparison because this sample size is commonly used to monitor PRRSV in North American breeding herds.
Aggregate samples used in monitoring infectious agents include bulk-tank milk samples, environmental swabs, air samples, or oral fluid samples.5-11 Overall, this is a highly cost-effective approach for improved monitoring. For example, the monthly cost of testing 30 piglet serum samples pooled by five is approximately 150 USD. Instead, the same 150 USD could be spent on six processing fluids per month representing hundreds of piglets. Alternatively, a 2500-sow herd producing an average of 1550 weaned piglets per week could test every piglet born in a week (approximately 1650 liveborn) by PRRSV rRT-PCR at a cost of approximately 100 USD (four processing fluids samples per week). The cost of testing the same number of pigs by PRRSV rRT-PCR in serum pooled by five would be approximately 8250 USD (330 PCRs at 25 USD each) per week.
It has been documented that PRRSV replicates in testicular epithelial cells and macrophages.13 Therefore, it makes biological plausibility that bodily fluids originated from castration and tail docking are suitable samples for PRRSV detection. At the piglet processing age (3 to 5 days old), PRRSV infection may have taken place in gestation (transplacental infection) or shortly after farrowing. Situations in which processing-fluid-based sampling can be used include monitoring breeding herds undergoing PRRSV elimination to determine whether there is virus circulation at the piglet processing-age group or to establish the optimum timing to intensify internal biosecurity practices (ie, no evidence of virus circulation in piglets being processed). Likewise, processing fluids offer an efficient method for continuous surveillance in breeding herds presumed to be PRRSV-negative. Perhaps most importantly, regional and national PRRSV elimination programs will benefit from this practical, simple, and affordable approach.
Regardless of the application, the design of monitoring and surveillance schemes will become more flexible and easily integrated with the daily routine due to the ease of implementation and lower costs associated with processing fluids. Whatever sampling design is ultimately implemented, testing more pigs more frequently will result in improved herd-level sensitivity for the detection of PRRSV and other pathogens. This may be a great tool for veterinarians to make informed interventions to decrease the time-to-detect PRRS outbreaks and increase the probability of detecting virus at near-zero prevalence. More studies are needed to further evaluate the herd sensitivity of processing fluids for PRRSV and other pathogen monitoring systems. This simple development promises to be a major breakthrough in disease monitoring and surveillance.
Implications
• Processing fluid is an aggregate sample easily obtained by farm staff under field conditions.
- The use of processing fluids makes it possible to test more pigs, more frequently for PRRSV.
- Processing fluids are a major improvement in disease surveillance systems and may increase the strength of PRRSV control and elimination programs.
Acknowledgments
This work was supported by Zoetis Inc. We thank Drs Pete Thomas, Deb Murray, Sara Dillon Hough, and Emily Byers for collaborating with this study by facilitating sample collection in breeding herds.
Conflict of interest
Dr Jose Angulo is employed by Zoetis Inc, that provided funding for the study.
Disclaimer
Scientific manuscripts published in the Journal of Swine Health and Production are peer reviewed. However, information on medications, feed, and management techniques may be specific to the research or commercial situation presented in the manuscript. It is the responsibility of the reader to use information responsibly and in accordance with the rules and regulations governing research or the practice of veterinary medicine in their country or region.
References
1. Holtkamp DJ, Kliebenstein JB, Neumann EJ, Zimmerman JJ, Rotto HF, Yoder TK, Wang C, Yeske PE, Mowrer CL, Haley CA. Assessment of the economic impact of porcine reproductive and respiratory syndrome on United States pork producers. J Swine Health Prod. 2013;21(2):72-84.
2. Corzo CA, Mondaca E, Wayne S, Torremorell M, Dee S, Davies P, Morrison RB. Control and elimination of porcine reproductive and respiratory syndrome virus. Virus Res. 2010;154(1-2):185-192.
3. Holtkamp D, Polson D, Torremorell M, Morrison R, Classen D, Becton L, Henry S, Rodibaugh MT, Rowland RR, Snelson H, Straw B, Yeske P, Zimmerman J. Terminology for classifying swine herds by porcine reproductive and respiratory syndrome virus status. J Swine Health Prod. 2011;19(1):44-56.
4. Linhares D. Evaluation of Immune Management Strategies to Control and Eliminate Porcine Reproductive and Respiratory Syndrome Virus (PRRSv) [dissertation]. Saint Paul: University of Minnesota; 2013.
5. Kittawornrat A, Panyasing Y, Goodell C, Wang C, Gauger P, Harmon K, Rauh R, Dufresne L, Levis I, Zimmerman J. Porcine reproductive and respiratory syndrome virus (PRRSV) surveillance using pre-weaning oral fluid samples detects circulation of wild-type PRRSV. Vet Microbiol. 2014;168(2-4):331-339.
6. Ramirez A, Wang C, Prickett JR, Pogranichniy R, Yoon KJ, Main R, Johnson JK, Rademacher C, Hoogland M, Hoffmann P, Kurtz A, Kurtz E, Zimmerman J. Efficient surveillance of pig populations using oral fluids. Prev Vet Med. 2012;104(3-4):292-300.
7. Prickett JR, Zimmerman JJ. The development of oral fluid-based diagnostics and applications in veterinary medicine. Anim Health Res Rev. 2010;11(2):207-216.
8. Prickett JR, Cutler S, Kinyon JM, Naberhaus N, Stensland WR, Yoon K-J, Zimmerman JJ. Stability of porcine reproductive and respiratory syndrome virus and antibody in swine oral fluid. J Swine Health Prod. 2010;18(4):187-195.
9. Prickett JR, Kim W, Simer R, Yoon K-J, Zimmerman J. Oral-fluid samples for surveillance of commercial growing pigs for porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 infections. J Swine Health Prod. 2008;16(2):86-91.
10. Prickett J, Simer R, Christopher-Hennings J, Yoon KJ, Evans RB, Zimmerman JJ. Detection of Porcine reproductive and respiratory syndrome virus infection in porcine oral fluid samples: a longitudinal study under experimental conditions. J Vet Diagn Invest. 2008;20(2):156-163.
11. Kittawornrat A, Prickett J, Chittick W, Wang C, Engle M, Johnson J, Patnayak D, Schwartz T, Whitney D, Olsen C, Schwartz K, Zimmerman J. Porcine reproductive and respiratory syndrome virus (PRRSV) in serum and oral fluid samples from individual boars: Will oral fluid replace serum for PRRSV surveillance? Virus Res. 2010;154(1-2):170-176.
12. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74(12):5463-5467.
13. Sur JH, Doster AR, Christian JS, Galeota JA, Wills RW, Zimmerman JJ, Osorio FA. Porcine reproductive and respiratory syndrome virus replicates in testicular germ cells, alters spermatogenesis, and induces germ cell death by apoptosis. J Virol. 1997;71(12):9170-9179.