| Brief communication | Peer reviewed |
Cite as: Qi X, Huang N, Zhao B, et al. Prevalence of serogroups and genotypes for fimbriae and enterotoxins in Escherichia coli isolated from diarrheic piglets in western China. J Swine Health Prod. 2012;20(6):290–293.
Also available as a PDF.
SummaryThe prevalence of O serogroups and virulence factors of Escherichia coli strains isolated from diarrheic pigs in western China were examined. Three uncommon O serogroups of E coli strains were discovered, and a high prevalence of enterotoxigenic isolates that produce only K99 or STa was detected. | ResumenSe examinó la prevalencia de serogrupos O y factores de virulencia de cepas aisladas de Escherichia coli de cerdos diarreicos en el oeste de China. Se descubrieron tres cepas poco comunes de serogrupos O de E coli, y se detectó una alta prevalencia de aislados enterotoxigénicos que producen solo K99 o STa. | ResuméLa prévalence des sérogroupes O et des facteurs de virulence de souches d’Escherichia coli isolées de porcelets diarrhéiques en Chine occidentale ont été étudiées. Trois sérogroupes O inhabituels de souches d’E coli ont été retrouvés, et une prévalence élevée d’isolats entérotoxinogènes ne produisant que K99 ou STa fut notée. |
Keywords: swine, enterotoxin, Escherichia coli diarrhea, fimbriae, O serogroups
Search the AASV web site
for pages with similar keywords.
Received: November 1, 2011
Accepted: May 30, 2012
Porcine neonatal diarrhea and postweaning diarrhea caused by enterotoxigenic Escherichia coli (ETEC) result in significant morbidity and mortality and are economically important diseases of pigs.1 Secretory diarrhea associated with ETEC infection is mediated by any of several enterotoxins, including heat labile enterotoxin (LT), heat-stable enterotoxin-a (STa), and heat-stable enterotoxin-b (STb).2 Enterotoxigenic E coli colonization in the small intestine is primarily mediated by fimbriae that attach to receptors on villus enterocytes.3 Fimbriae, including K88 (F4), K99 (F5), 987P (F6), F18, and F41, are commonly found in porcine E coli strains, and ETEC strains expressing these fimbriae are clinically important in porcine diarrhea.4
Identification of fimbriae and enterotoxin prevalence by geographic location is an important epidemiological tool for the control of colibacillosis in young pigs.5 The purpose of the present study was to investigate the prevalence of various O serogroups and virulence factors of E coli strains isolated from diarrheic piglets in western China.
Materials and methods
Selection of study farms
Three to four swine-production facilities were chosen from each of the five provinces (Shaanxi, Gansu, Ningxi, Sichuan, and Yunnan) located in western China, where the climate is dry. Sixteen farms were selected according to the following criteria: neonatal diarrhea and postweaning diarrhea were recognized problems, the herd included more than 500 sows, the farms were 500 to 800 km apart, and the farm’s owner and staff were willing to participate in the study. All sows and litters were housed indoors and fed a commercial feed ad libitum. Electric heat lamps provided supplemental heat. Sows and pigs were purebred or crossbred Duroc, Large White, or Landrace.
Bacterial isolates
Two hundred and eight E coli field isolates (103 from piglets with neonatal diarrhea and 105 from pigs with postweaning diarrhea) were collected between 2008 and 2010. A case of diarrhea was recorded when a pig was observed to pass liquid feces and showed signs of dehydration at 1 to 50 days of age. A litter was recorded as being affected by diarrhea when it occurred in > 30% of piglets in the litter. A total of 235 rectal swabs were collected from 1- to 50-day-old diarrheic pigs. One swab was collected from each diarrheic pig, with a total of 10 to 15 diarrheic samples collected from each farm. The swabs were cultured directly on MacConkey agar, and suspect E coli isolates were identified using biochemical procedures.6 Isolates were stored frozen at -70°C and were not subcultured more than twice before further investigation. Escherichia coli strains obtained from the China Institute of Veterinary Drug Control (Beijing, China) were used as controls and included C83903 (K88:STa:STb), C83917 (987P), C83914 (K99:LT:STb), 107/86 (F18), and C83921 (F41).
O serogrouping
Serotyping was performed by standard methods at the China Institute of Veterinary Drug Control.1 Escherichia coli isolates were grown overnight in tryptic soy broth and then autoclaved for 2 hours. A microtiter plate-agglutination test was initially performed on all strains against a total of 171 O group antisera in predetermined dilutions, and positive reactions were confirmed by a tube agglutination test.
Hemolysis determination
Escherichia coli isolates were cultured on agar plates (supplemented with 5% fetal calf blood) for 18 hours at 37°C, and the hemolytic activity of each strain was visually determined.
Polymerase chain reaction assay
All isolates were examined for genes encoding fimbriae and enterotoxins. Table 1 lists the base sequences and predicted sizes of amplified products for the specific oligonucleotide primers used in the present study. All polymerase chain reaction (PCR) assays for fimbriae and enterotoxins were performed as previously described with modifications.7 A 50-μL PCR reaction included 5 μL bacterial deoxyribonucleic acid (DNA) template, 5 μL 10× DNA polymerase buffer, 5 μL 25 mM MgCl2, 20 mM each deoxyribonucleoside triphosphate, 0.5 U Taq DNA polymerase in storage buffer A, and 1 μL of each primer appropriate for the assay. The composition of storage buffer A was 50 mM Tris-HCl (pH 8.0 at 25°C), 100 mM NaCl, 0.1 mM ethylenediaminetetra-acetic acid (EDTA), 1 mM dithiothreitol, 50% glycerol, and 1% Triton X-100. Reactions were subjected to one cycle at 94°C for 5 minutes, followed by 30 cycles at 94°C for 45 seconds, 54°C for 45 seconds, and 72°C for 45 seconds, and a final extension at 72°C for 10 minutes.
Table 1: Polymerase chain reaction primers used to amplify virulence genes of Escherichia coli isolates*
* Two hundred and eight E coli field isolates were collected between 2008 and 2010 from 235 rectal-swab samples from 1- to 50-day-old diarrheic pigs on 16 farms located in western China. † The method of Zhang et al7 was used for LT, STa, STb, and Stx2e and the method of Chen et al1 for K88, K99, 987P, F41, and F18. |
The amplified products were visualized by standard gel electrophoresis of 5 μL of the final reaction mixture in 1.5% agarose gel in Tris-acetate-EDTA buffer, performed at 5 V per cm. The gels were stained with ethidium bromide (5 μg per mL), visualized under ultraviolet light (300 nm), and photographed with a camera system. A 100-bp DNA ladder (Sigma-Aldrich, Inc, St Louis, Missouri) was used as molecular-size marker (100 to 2000 bp). Polymerase chain reaction was performed in triplicate for each sample.
Detection of shiga toxin by western blot assay
A western blot assay, described previously,8 was used to detect shiga toxin 2e (Stx2e). The E coli isolates that carried the shiga-toxin gene, as determined by PCR assay, were inoculated into 2× YT medium (Sigma-Aldrich, Inc) supplemented with ampicillin (100 µg per mL) and grown at 37°C with agitation until an OD600 of 0.4 to 0.6 was reached. The bacterial pellets were obtained by centrifugation at 5000g for 10 minutes at 4°C. Following lysis and sonication, the Stx2e from E coli was purified by nickel ion affinity chromatography using a HisTrap Kit (Amersham Pharmacia, Inc, Piscataway, New Jersey) according to the instructions of the manufacturer. Western blots were performed with sodium dodecylsulfate-15% polyacrylamide gel electrophoresis slab gels. After transfer onto polyvinylidene difluoride membranes (Millipore, Bedford, Massachusetts), filters were blocked with 5% nonfat milk and then incubated with polyclonal antibodies raised in rabbits to Stx2e (kindly provided by the National Institute for Epidemic Disease, Beijing, China), followed by a goat anti-rabbit alkaline phosphatase-conjugated antibody (Sino-American Bio Co, Tianjin, China). Protein bands were visualized using an enhanced chemiluminescence protocol Western Blotting Detection Kit (Amersham Biosciences).
Results
O serogroups
Table 2 illustrates the distribution of serogroups among the 208 isolates. Of these isolates, 166 were serotyped, 29 could not be serotyped, and 13 auto-agglutinated. Although the isolates were distributed among 13 serogroups, 132 (63% of the 208 isolates) belonged to seven serogroups, namely, O80, O141, O139, O6, O9, O20, and O101. Escherichia coli O80 and O141 strains comprised approximately 24% and 14% of all isolates, respectively, whereas strains belonging to other O groups were markedly less prevalent, ranging from approximately 1% to 7% (Table 2). Seven isolates were E coli O157 strains.
Table 2: Serogroups and hemolytic activity found in 208 Escherichia coli isolates from diarrheic piglets*
* Source of samples described in Table 1. Escherichia coli isolates were cultured on blood agar plates, and the hemolytic activity of each strain was visually determined. NT = nontypeable; OR = O-rough. |
Hemolysis determination
Among the 208 E coli isolates, 65 and 84 strains were alpha-hemolytic and beta-hemolytic, respectively (Table 2). Hemolytic isolates dominated in serogroups O80 (31 of 49) and O141 (26 of 29), as well as in the NT isolate group (26 of 29).
Fimbrial antigen, serogroup, and hemolytic activity
The prevalence of the K88, K99, F41, 987P, and F18 genes in the 208 E coli isolates were examined in this study. Over two thirds of the isolates (70%) were positive for one fimbrial gene (132 isolates) or more (14 isolates). Among the isolates positive for at least one fimbrial gene, 88 were K99-positive (76 strains isolated from 1- to 3-week old diarrheic piglets and 12 strains isolated from 3- to 7-week old pigs with postweaning diarrhea), whereas only one was F41-positive. The F18, K88, K88+987P, 987P, and K88+F18 genes were detected in 21, 14, 12, eight, and two isolates, respectively. Isolates that carried the fimbrial genes were widespread among the serogroups. One hundred and thirty of the 149 hemolytic isolates harbored one or more fimbrial genes.
Toxin genes
A total of 112 E coli isolates were positive for STa (25%), LT (9%), LT and STa (10%), STb (7%), or STa and STb (3%) genes. The Stx2e gene was detected by PCR in five of seven E coli O157 strains. A representative western blot is shown in Figure 1, demonstrating the A and B subunits of Stx2e carried by E coli isolates. Ninety-one isolates (44%) did not harbor any of the toxin genes investigated.
Figure 1: A representative western blot analysis of the Shiga toxin 2e (Stx2e) expressed by an Escherichia coli strain in which the Stx2e gene was detected by polymerase chain reaction assay. This E coli strain was isolated from diarrheic pigs in western China. Western blots were performed using polyclonal antibody against Stx2e obtained from the National Institute for Epidemic Disease (Beijing, China). Lane 1: DNA Marker; Lane 2: A (32 kDa), A1 (27.5 kDa), and B (7.5 kDa) subunits of Stx2e protein. 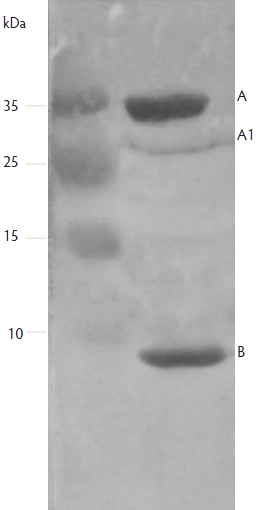 |
Discussion
While many different E coli serogroups have been identified as the one of primary importance in causing diarrhea in piglets,9 the distribution and frequencies of these serogroups can vary considerably from region to region and over time in a given region.10 In the present study, some novel serogroups were identified, specifically serogroups O80, O6, and O38. This finding implies that regional differences or other selective pressure allowed E coli from these O serogroups to proliferate in the intestines of pigs from the surveyed western regions in China.
Enterotoxigenic E coli strains that produce K88 (F4) or F18 fimbrial genes are commonly associated with postweaning diarrhea in young pigs.1 The present study confirmed that K88-positive and F18-positive isolates are the predominant fimbrial ETEC strains associated with postweaning diarrhea in pigs in western China. In addition, K99 (F5), 987P (F6), or F41 fimbrial antigens are most frequently encountered in ETEC from pigs with neonatal diarrhea.11 These findings are in agreement with the present results, in which K99 colonization factors were identified in 76 of 103 piglets younger than 3 weeks. A previous study reported that the K99 antigen is more prevalent in the dry season than in the rainy season in Central Vietnam.12 This condition implies that the dry climate in western China may have also contributed to the high prevalence of K99 fimbriae in the present study.
The pattern of enterotoxins produced by ETEC strains varies among countries and regions, and conflicting results have been published.8 Consistent with other studies,1,6 STa- and LT-producing strains were more prevalent than STb-producing strains in the present study. Shiga-toxin-producing E coli have recently emerged as important food-borne pathogens, especially serotype O157:H7.13 In the present study, five of seven E coli O157 strains carried the Shiga-toxin gene. These data confirm previous reports that pigs are a potential reservoir for E coli O157:H7 and a risk to human health.8
Implications
• Serological surveillance for E coli subtypes in piglets in western China is essential to public health because of the occurrence of strains of E coli belonging to subgroup O157, which cause severe disease in humans worldwide.
• It is essential to take into consideration the age-related phenotypic characteristics of ETEC in order to produce effective vaccines against enteric infections of piglets in western China.
Acknowledgements
This work was financially supported by the Foundation for The Excellent Youth Scholars and Key Teacher of Northwest A & F University and Excellent Researcher Special Funds of Northwest A & F University.
References
1. Chen X, Gao S, Jiao X. Prevalence of serogroups and virulence factors of Escherichia coli strains isolated from pigs with postweaning diarrhoea in Western China. Vet Microbiol. 2004;103:13–20.
2. Zhang W, Berberov EM, Freeling J, He D, Moxley RA, Francis DH. Significance of heat-stable and heat-labile enterotoxins in porcine colibacillosis in an additive model for pathogenicity studies. Infect Immun. 2006;74:3107–3114.
3. Fleckenstein JM, Hardwidge PR, Munson GP, Rasko DA, Sommerfelt H, Steinsland H. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect. 2010;12:89–98.
4. Madoroba E, Van Driessche E, De Greve H, Mast J, Ncube I, Read J, Beeckmans S. Prevalence of enterotoxigenic Escherichia coli virulence genes from scouring piglets in Zimbabwe. Trop Anim Health Prod. 2009;41:1539–1547.
5. Cheng DR, Sun HC, Xu J, Gao S. PCR detection of virulence factor genes in Escherichia coli isolates from weaned piglets with edema disease and/or diarrhea in China. Vet Microbiol. 2006;115:320–328.
6. Kim YJ, Kim JH, Hur J, Lee JH. Isolation of Escherichia coli from piglets in South Korea with diarrhea and characteristics of the virulence genes. Can J Vet Res. 2010;74:59–64.
7. Zhang W, Zhao M, Ruesch L, Omot L, Francis D. Prevalence of virulence genes in Escherichia coli strains recently isolated from young pigs with diarrhea in the US. Vet Microbiol. 2007;123:145–152.
8. Sánchez S, Martinez R, Garcia A, Vidal D, Blanco J, Blanco M. Detection and characterization of O157:H7 and non-O157 Shiga toxin-producing Escherichia coli in wild boars. Vet Microbiol. 2010;14:420–423.
9. Zhang W, Robertson DC, Zhang C, Bai W, Zhao M, Francis DH. Escherichia coli constructs expressing human or porcine enterotoxins induce identical diarrheal diseases in a piglet infection model. Appl Environ Microbiol. 2008;74:5832–5837.
10. Vu-Khac H, Holoda E, Pilipcinec E, Blanco M, Blanco JE, Dahbi G. Serotypes, virulence genes, intimin types and PFGE profiles of Escherichia coli isolated from piglets with diarrhea in Slovakia. Vet J. 2007;174:176–187.
*11. Francis DH. Diagnostic notes. Enterotoxigenic Escherichia coli infection in pigs and its diagnosis. J Swine Health Prod. 2002;10:171–175.
12. Hong TT, Linh NQ, Ogle B, Lindberg JE. Survey on the prevalence of diarrhoea in pre-weaning piglets and on feeding systems as contributing risk factors in smallholdings in Central Vietnam. Trop Anim Health Prod. 2006;38:397–405.
13. Rotariu O, Ogden ID, Macritchie L, Forbes KJ, Williams AP, Cross P, Hunter CJ, Teunis PFM, Strachan NJC. Combining risk assessment and epidemiological risk factors to elucidate the sources of human E coli O157 infection. Epidemiol Infect. 2011;27:1–16.
* Non-refereed reference.