Grau-Roma L, Baekbo P, Rose N, et al. Clinical and laboratory studies on herds affected with postweaning multisystemic wasting syndrome in Denmark, France, Spain, and Sweden: Disease progression and a proposal for herd case definition. J Swine Health Prod. 2012;20(3):129–136
| Original research | Peer reviewed |
Cite as: Grau-Roma L, Baekbo P, Rose N, et al. Clinical and laboratory studies on herds affected with postweaning multisystemic wasting syndrome in Denmark, France, Spain, and Sweden: Disease progression and a proposal for herd case definition. J Swine Health Prod. 2012;20(3):129–136.
Also available as a PDF.
SummaryObjectives: To propose and evaluate a protocol to establish a diagnosis of postweaning multisystemic wasting syndrome (PMWS) at herd level. Materials and methods: The data used included both laboratory data from previous epidemiological studies carried out in Italy, Denmark, and Spain and original postweaning mortality data collected from several herds in Denmark, France, Spain, and Sweden. Statistical process control techniques were used to analyze the longitudinal evolution of mortality in each herd. Results: The analysis of data sets from three different countries showed that to diagnose at least one PMWS-affected pig with a probability > 95%, it is necessary to study a minimum of three to five pigs. Longitudinally collected data showed that when > 20 data points were available, a significant increase in mortality was always detected at close to the time of PMWS diagnosis. When < 10 points were available, a significant increase in mortality was detected in four of six studied herds, although mortality percentages were always higher at the time of PMWS diagnosis than before diagnosis. Implications: These results suggest that two conditions must be fulfilled to diagnose PMWS in a herd. Firstly, a significant increase in postweaning mortality, compared to the historical background in the herd, must be observed in association with clinical signs compatible with PMWS. Secondly, PMWS must be diagnosed in at least one of three to five necropsied pigs concurrently with the increase in mortality. Ruling out other potential causes of increased mortality is also necessary. | ResumenObjetivos: Proponer y evaluar un protocolo para establecer un diagnóstico del síndrome multisistémico de desmedro post destete (PMWS por sus siglas en inglés) a nivel de hato. Materiales y métodos: Los datos utilizados incluyeron datos de laboratorio procedentes de estudios epidemiológicos previos realizados en Italia, Dinamarca, y España así como datos originales de mortalidad post-destete procedentes de distintas granjas de Dinamarca, Francia, España, y Suecia. Se utilizaron técnicas de control estadístico del proceso para analizar la evolución longitudinal de la mortalidad en cada hato. Resultados: El análisis de la información de tres países diferentes mostró que para diagnosticar al menos un cerdo afectado por el PMWS con una probabilidad > 95%, es necesario estudiar un mínimo de tres a cinco cerdos. La información recolectada longitudinalmente mostró que cuando hubo > 20 puntos de información disponible, siempre se detectó un incremento significativo en la mortalidad cerca del momento del diagnóstico del PMWS. Cuando hubo < 10 puntos disponibles, se detectó un incremento significativo en la mortalidad en cuatro de los seis hatos estudiados, aunque los porcentajes de mortalidad siempre fueron más altos al momento del diagnóstico del PMWS que antes del diagnóstico. Implicaciones: Estos resultados sugieren que deben cumplirse dos condiciones para diagnosticar el PMWS en un hato. Primero, debe observarse un incremento significativo, comparado con el antecedente histórico del hato, en la mortalidad post destete asociado a signos clínicos compatibles con el PMWS. Segundo, debe haberse diagnosticado el PMWS en al menos una de las tres a cinco necropsias efectuadas en los cerdos simultáneamente con el incremento en la mortalidad. También es necesario descartar otras causas potenciales en el incremento en la mortalidad. | ResuméObjectifs: Proposer et évaluer un protocole pour établir un diagnostic à l’échelle du troupeau du syndrome multi-systémique de dépérissement en post-sevrage (PMWS). Matériels et méthodes: Les données utilisées incluaient conjointement les données de laboratoire d’études épidémiologiques antérieures effectuées en Italie, au Danemark, et en Espagne, ainsi que des données originales de mortalité post-sevrage amassées dans plusieurs troupeaux au Danemark, en France, en Espagne, et en Suède. Des techniques de maîtrise statistique des procédés ont été utilisées pour analyser l’évolution longitudinale de la mortalité dans chaque troupeau. Résultats: L’analyse des regroupements de données provenant de trois pays différents a démontré que pour diagnostiquer au moins un porc affecté par PMWS avec une probabilité supérieure à 95%, il est nécessaire d’étudier un minimum de trois à cinq porcs. Les données recueillis longitudinalement ont montré que lorsque plus de 20 données sont disponibles, une augmentation significative de la mortalité était toujours détectée près du moment du diagnostic de PMWS. Lorsque moins de 10 données étaient disponibles, une augmentation significative de la mortalité était détectée dans quatre des six troupeaux étudiés, bien que les pourcentages de mortalité fussent toujours plus élevés au moment du diagnostic de PMWS qu’avant le diagnostic. Implications: Les résultats obtenus suggèrent que deux conditions doivent être remplies pour poser un diagnostic de PMWS dans un troupeau. Premièrement, une augmentation significative de la mortalité post-sevrage, comparativement aux données de base historiques dans le troupeau, doit être observée en association avec des signes cliniques compatibles avec PMWS. Deuxièmement, un diagnostic de PMWS doit être posé pour au moins un des trois à cinq porcs soumis pour nécropsie au moment de l’augmentation de la mortalité. L’élimination des autres causes potentielles d’augmentation de mortalité est également nécessaire. |
Keywords: swine, postweaning multisystemic wasting syndrome, porcine circovirus type 2, diagnosis, statistical process control, PMWS, PCV2
Search the AASV web site
for pages with similar keywords.
Received: January 11, 2011
Accepted: January 6, 2012
Postweaning multisystemic wasting syndrome (PMWS) is a multifactorial pig disease in which porcine circovirus type 2 (PCV2) is the essential infectious agent.1 Among the conditions included within the scope of porcine circovirus diseases (PCVDs) or porcine circovirus associated disease (PCVAD), PMWS has major clinical and economic impacts.1,2 This disease most commonly affects pigs from 2 to 4 months of age, ie, in the late nursery and growing phases.3-5 Morbidity and lethality rates are variable depending on the herd and the batch of animals, ranging most commonly from 4% to 30% and 70% to 80%, respectively, with lethality rate defined as the percentage of pigs that die among those showing morbidity.5 However, cases have also been described where morbidity is exceptionally high (> 50%) or where a few pigs are sporadically affected in herds with very good production records.5 The most common clinical sign is wasting, although affected pigs also frequently show other signs, such as fever, skin pallor, dyspnea, diarrhea, and non-response to antimicrobial treatment.3-6 Despite this, affected pigs are often medically treated, with a resultant increase in antimicrobial usage.7
Wasting and respiratory signs in a proportion of late nursery and growing pigs, together with the presence of lesions such as enlarged lymph nodes, interlobular pulmonary edema, or white-spotted kidneys, are suggestive of PMWS. However, such gross lesions are not always observed and clinical signs alone are nonspecific. Therefore, none of them are sufficient to establish the diagnosis with certainty. Infection with PCV2 occurs in virtually all herds worldwide.8 Thus, laboratory results must be interpreted with caution.9,10 Today, an individual pig is considered PMWS affected when the three following criteria are fulfilled: first, presence of compatible clinical signs, including wasting or growth retardation; second, observation of characteristic histopathological lesions in lymphoid tissues, including lymphocyte depletion and histiocytic infiltration; and third, detection of moderate to large amounts of PCV2 within the lesions in lymphoid and other tissues of affected pigs (using either immunohistochemistry [IHC] or in situ hybridization [ISH] techniques).1,11 Taking into account the strong correlation observed between the amount of PCV2 antigen or nucleic acid and the severity of PMWS histopathological lesions, disease definition implies also moderate to severe histopathological lesions and a moderate to large amount of PCV2 detected.5 Techniques such as polymerase chain reaction (PCR) or serology lack adequate sensitivity and specificity to diagnose PMWS.10,12
The relatively recent introduction of commercial PCV2 vaccines has dramatically changed the approach to PMWS prevention. Such vaccines have become excellent tools to prevent and diminish the impact of PMWS.13-16 However, the first step before implementing any vaccination program should be to ensure that the herd is affected by the corresponding disease. Establishing diagnosis of the disease prevents a potential “vaccine failure” perception and saves time and money. Up to the present time, a herd has been considered clinically affected by PMWS when high percentages of wasting and mortality occur postweaning (ie, in the nursery and growing and finishing areas) and individual pigs fulfill requirements for diagnosis of PMWS.1 However, a formal study on the criteria to establish disease diagnosis at herd level has not yet been performed. For these reasons, and with the experience of > 5 years of diagnostic and investigation experience by a consortium of 15 European and American research groups (www.pcvd.eu), the present study was designed to explore and propose a potential scientific-technical herd case definition of epidemic PMWS by retrospectively assessing clinical and laboratory data from herds in which the disease had already been diagnosed in individual animals. The second objective was to provide a technical approach to objectively quantify and assess whether or not a given herd is suffering a significant impact due to PMWS. The proposed technical definition was explored on the basis of two elements: firstly, clinical appearance in the herd (ie, pigs showing clinical signs compatible with PMWS, quantified numerically by considering combined mortality in the nursery and growing-finishing phases) and, secondly, laboratory examination of necropsied pigs fulfilling the internationally accepted criteria for diagnosis of PMWS in individual animals.1,11
Data was analyzed using statistical process control (SPC) techniques, an analytic approach that “explains with statistical confidence when a process performance is improving, staying the same, or getting worse.”17 These techniques are increasingly applied by livestock herd managers and their consultants and by scientists working with longitudinal data from livestock.17 Specifically, control charts, which allow identifying and distinguishing between normal (common cause) and abnormal (special cause) variability, have already been reported in swine production.18,19 When only common-cause variation is present in process output, the process is said to be operating under a state of statistical control. The process enters an out-of-control state when an aspect or some aspects of the process change and impact process performance.
Materials and methods
Herds and animals
To determine the sample size adequate to establish a PMWS diagnosis in individual pigs, data from two previously published studies were used.12,20 Those studies were performed in a total of 56 PMWS-affected herds in Denmark (n = 8),12 Spain (n = 3),12 and Italy (n = 45).20 A total of 330 wasted pigs (91 from Denmark, 79 from Spain, and 160 from Italy) were necropsied and lymphoid tissues were examined histopathologically and by IHC or ISH for PCV2 detection.
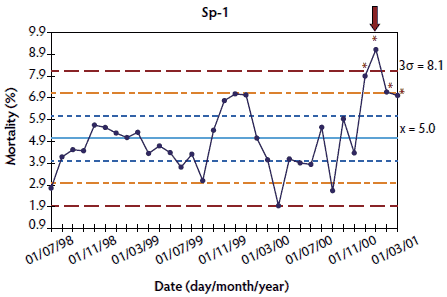
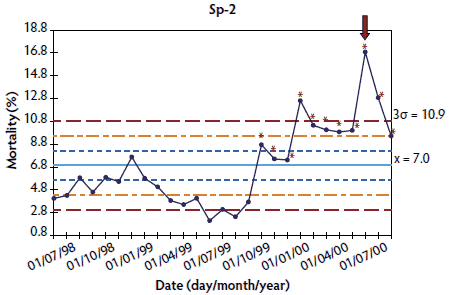
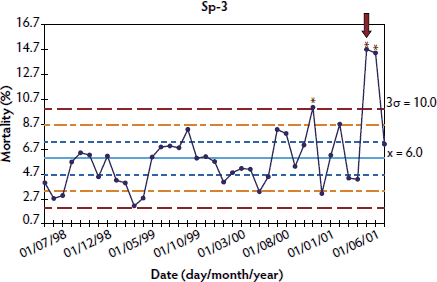
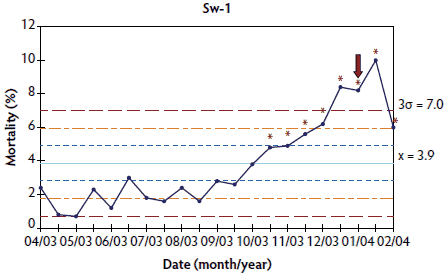
In order to analyze the evolution of mortality before and after a PMWS outbreak, three different herds per country in Denmark (Dk-1, Dk-2, and Dk-3), France (Fr-1, Fr-2, and Fr-3), and Spain (Sp-1, Sp-2, and Sp-3), and one in Sweden (Sw-1),21 were retrospectively selected, taking into account the fulfillment of two conditions: firstly, availability of a PMWS diagnosis in at least one pig; and, secondly, longitudinal mortality data available during a period before PMWS occurred (between 9 and 35 months, depending on the herd) and during the time when PMWS was occurring. Table 1 summarizes the herds used for performing each of these analyses and the country of origin of each herd.
Table 1: Farms used to determine the sample size needed to establish a diagnosis of postweaning multisystemic wasting syndrome (PMWS) and to analyse the longitudinal evolution of mortality percentages
|
Parameters evaluated and statistical analyses
The proportion of pigs showing clinical signs compatible with PMWS that fulfilled the criteria for PMWS individual diagnosis obtained in previous studies was used to calculate the probability of diagnosing the disease in at least one animal, depending on the number of necropsied pigs.12,20 The calculation was made by means of a theoretical approach as described elsewhere,22,23 applying the formulas detailed below:
P = (1 – Pr)X
P´= 1 – P
where P is the probability of not diagnosing the disease in any pig, Pr is the proportion of pigs diagnosed with PMWS of the total number of necropsied (suspected) pigs, X is the number of necropsied pigs, and P´ is the probability of diagnosing the disease in at least one pig.
Mortality data was collected monthly in Spanish herds, at 2-week intervals in the Swedish herd, and at 3- to 6-month intervals in French and Danish herds, including both nursery and growing-finishing phase mortality. Data represent the means of percentage mortality in the given periods of time. Data was analyzed using QIMacros2007 SPC for Excel (KnowWare International Inc; www.excel-spc-software.com/excel-spc-software.html). Control charts (X-charts) were created by plotting longitudinal mortality measurements consecutively collected in each herd prior to PMWS diagnosis. Overall average (central line) and upper and lower control limits (corresponding to + 3σ [standard deviation] and – 3σ) provide a visual means for understanding the amount of variation inherent in a system over time. When data points appear within the control limits without any unusual patterns, then the process is considered to exhibit common cause of variation. Therefore, it is considered to be in statistical control (or stable). System changes were considered significant if one or several of the following four conditions existed: one single point more than 3σ above the mean; at least two of three successive points 2σ above the mean; at least four of five successive points 1σ above the mean; or nine or more successive points above the mean.17,24 These are the conditions generally recommended when using SPC techniques, with the only difference being that variations are usually considered when they occur either above or below the mean.17,24 However, a substantial decrease in mortality percentage is beneficial and, consequently, cannot be considered an alarm in a disease context. Thus, in the present proposal, the general conditions have been applied only for an increase in mortality.
Results
The percentages of suspected pigs that fulfilled PMWS diagnosis in Italy, Denmark, and Spain were 69%,20 56%,12 and 46%,12 respectively. The probability of diagnosing at least one PMWS-affected pig in an affected herd, taking into account the data reported in those studies, is presented in Table 2. Results showed that to diagnose at least one PMWS-affected pig with a probability > 95%, it is necessary to study a minimum of three, four, and five pigs, considering Italian, Danish, and Spanish data, respectively.
Table 2: Probability of diagnosing one pig affected with postweaning multisystemic wasting syndrome depending on the number of necropsied pigs*
* Shading indicates probability > 95%. |
||||||||||||||||||||||||||||||||||||||||||||||||||
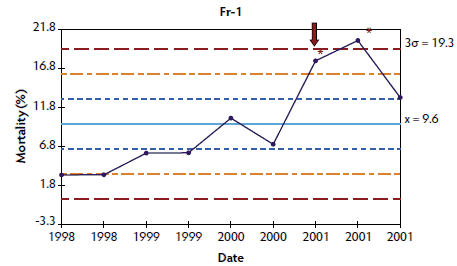
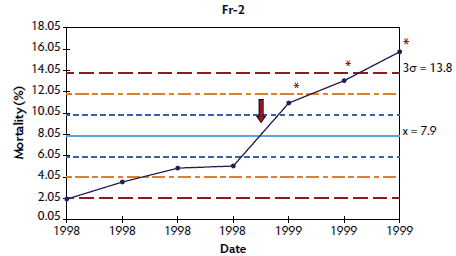
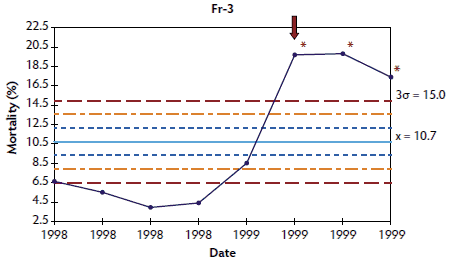
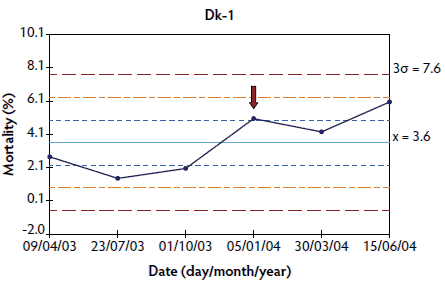
Longitudinal mortality data compiled from all longitudinally analyzed herds are presented as X-charts in Figures 1 through 4. Mortality data from Spanish and Swedish herds, for which > 20 data points were available, are presented in Figures 1 and 2, respectively. For Danish and French herds, in which < 10 points were available, mortality data are presented in Figures 3 and 4, respectively.
In herds where > 20 points were available, results showed that the system was out of control at the time of PMWS diagnosis (Figures 1 and 2). Specifically, mortality percentage at PMWS diagnosis was always higher than the upper control limit. Moreover, in herds Sp-1, Sp-2, and Sw-1, a statistically significant increase in mortality was already detected at one or more points before PMWS diagnosis. In Sp-1, a statistically significant increase in mortality was detected only in the point immediately before PMWS diagnosis. However, in Sp-2 and Sw-1, the statistically significant increase in mortality percentage was detected eight and five points before PMWS diagnosis, respectively. In addition, in herd Sp-3, a sporadic increase in mortality was detected six points before PMWS diagnosis.
Figure 1: Control charts of mortality data from three Spanish herds (Sp-1, Sp-2, and Sp-3) collected during an approximately 2-year period. The time when postweaning multisystemic wasting syndrome was diagnosed is indicated by a red arrow. Points where the system is unstable are indicated by red asterisks. The central light blue line represents the mean of mortality. Broken lines indicate upper and lower control limits (corresponding to ± 1σ, ± 2σ, and ± 3σ).
|
Figure 2: Control chart of mortality data from a Swedish herd (Sw-1) collected at 2-week intervals during an approximately 1-year period. The time when postweaning multisystemic wasting syndrome was diagnosed is indicated by a red arrow. Points where the system is unstable are indicated with red asterisks. The central light blue line represents the mean of mortality. Broken lines indicate upper and lower control limits (corresponding to ± 1σ, ± 2σ, and ± 3σ).
|
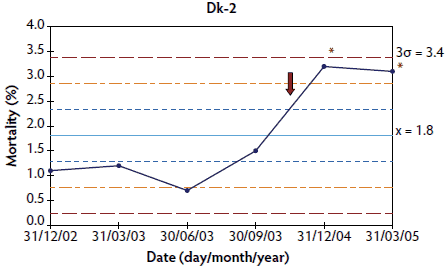
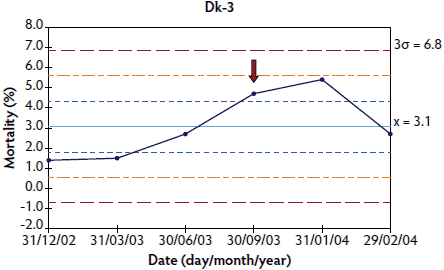
In herds in which < 10 points were available at the time of PMWS diagnosis, the system was out of control in four of the six studied herds: the three French herds (Figure 3) and one of the Danish herds (Figure 4). In herds Dk-1 and Dk-3, although mortality percentages at PMWS diagnosis were always higher than the values recorded before diagnosis, no changes in the stability of the system were detected, according to the statistical process control criteria. Furthermore, mortality percentage at the time of PMWS diagnosis was higher than the upper control limit in only one of the six studied herds (Fr-3).
Figure 3: Control charts of mortality data from three French herds (Fr-1, Fr-2, and Fr-3) collected during an approximately 1- to 3-year period. The time when postweaning multisystemic wasting syndrome was diagnosed in each herd is indicated by a red arrow. Points where the system is unstable are indicated with red asterisks. The central light blue line represents the mean of mortality. Broken lines indicate upper and lower control limits (corresponding to ± 1σ, ± 2σ, and ± 3σ).
|
Figure 4: Control charts of mortality data from three Danish herds (Dk-1, Dk-2, and Dk-3) collected during an approximately 1- to 3-year period. The time when postweaning multisystemic wasting syndrome was diagnosed in each herd is indicated by a red arrow. Points where the system is unstable are indicated with red asterisks. The central light blue line represents the mean of mortality. Broken lines indicate upper and lower control limits (corresponding to ± 1σ, ± 2σ, and ± 3σ).
|
Apart from PMWS, no other clinically significant diseases or pathogens associated with the observed increases in mortality were reported in the studied herds.
Discussion
Currently, criteria to establish a PMWS diagnosis in individual pigs are well defined and internationally accepted.1,11 However, to our knowledge, criteria to establish a PMWS diagnosis at herd level have not been reported in peer-reviewed journals, although guidelines have been suggested and published in Web pages.25 In the present work, a definition for PMWS herd diagnosis in herds experiencing epidemic PMWS is explored and proposed on the basis of scientific and technically sound data.
The present proposal for PMWS herd diagnosis is primarily based on overall clinical signs in terms of mortality and statistical calculations. Although mortality is a frequent clinical outcome of PMWS,1 the specificity of this definition relies on PMWS diagnosis in individual pigs. Thus, the first step to diagnose PMWS in a given herd should be individual PMWS definition in at least one pig. Results obtained from previous studies, where sick pigs from PMWS-affected herds were necropsied and histopathologically studied,12,20 showed that necropsies must be performed in at least three to five pigs showing clinical signs compatible with PMWS to be certain of diagnosing PMWS in at least one pig with a probability > 95%. The slight differences observed between the analyses of Spanish, Danish, and Italian data may have important implications for selecting the most appropriate pigs for necropsies. Lesions and viral load diminish with time,2 and it has been suggested that pigs should be selected to assess diagnostic criteria during the first week that they demonstrate clinical signs compatible with PMWS.26 In addition, differences between techniques and pathologists assessing histopathological lesions in laboratories may also contribute to differences obtained among countries.27 From a practical point of view, examining three to five pigs is usually affordable for practitioners in terms of time and physical effort.
It is important to take into account that, in herds with very good performance data, individual animals may fulfill the three individual diagnostic criteria for PMWS.28 However, when a herd is affected by PMWS, there is a worsening in general health status, causing increases in parameters such as mortality, number of culls, and feed conversion ratios, and decreases in other parameters, such as average daily gain.5,6 Therefore, a PMWS herd diagnosis should parallel a clinical problem present and detectable in the herd. Among the productive parameters affected by PMWS, mortality was the one chosen to explore a potential herd case definition for several reasons. This disease causes high mortality, especially in acute outbreaks,5,6 and the number of dead pigs is usually an available parameter,29 since almost all herds count the number of pigs entering and leaving a facility. Other production parameters, such as body weight, which is necessary to calculate average daily gain, are less easily attained, and they are therefore less likely to be available than mortality.
Statistical process control allows an investigator to explain with statistical confidence whether performance is remaining unchanged or getting better or worse with time.17 When SPC techniques were applied to longitudinal mortality data for PMWS herd diagnosis, it was observed that, in most studied herds (eight of 10), the system became unstable before or around the time when PMWS was diagnosed in individual animals. These results suggested that PMWS was directly related to an increase in mortality in the herds, and therefore it is a parameter that can potentially be used to establish a PMWS herd diagnosis.
It is stated that 20 to 25 data points are needed on a control chart before the limits are sufficiently “firmed-up,” allowing confident detection of a special-cause variation.24,30 With fewer data points, the resulting control limits are regarded as being “soft” or “provisional.”24 Indeed, mortality was not unstable according to SPC at the time of individual PMWS diagnosis in two of the 10 studied herds (Dk-1 and Dk-3) where only six data points were available. Nevertheless, although < 20 points were available for herds Dk-2, Fr-1, Fr-2, and Fr-3, an increase in mortality was detected at the time of PMWS diagnosis in these herds. This result supports the idea that control charts can be used even when few data points are available, although it is necessary to use > 20 points to avoid false-negative results.24 Instability in the system was detected at several points before individual PMWS diagnosis in herds Sp-2 and Sw-1, suggesting that PMWS was already present some weeks or months before the diagnosis was established by laboratory testing in these herds. Thus, there may be a delay between an increase in mortality and the decision to perform diagnostic studies, including histopathological examinations. However, it cannot be ruled out that the increases in mortality detected before PMWS diagnosis were at least partially due to other undetected diseases or pathogens.
Statistical process control is an accessible technique that allows producers to monitor the longitudinal evolution of mortality and provides an alarm signal when there is a disturbance in the normal behavior of the herd. The application of SPC technique, together with observation of clinical signs compatible with PMWS, will suggest to the veterinarian the necessity of performing necropsies and histopathological diagnosis. Thus, SPC techniques performed on mortality data represent an unbiased way to define a PMWS diagnosis on a herd basis.
Another approach might be the use of chi-square tests to detect a statistically significant increase in mortality by comparing proportions of mortality in different batches of pigs (during and before the PMWS outbreak). However, the chi-square test compares mortality rates at only two time points rather than providing an overview of the longitudinal evolution of mortality as SPC techniques do. Even if mortality records for several age categories are missing in a given herd, it is possible to define a significant increase in mortality when it exceeds the national or regional mean by 50%, as previously suggested in the framework of the European Consortium of 15 research groups on PCVD.25
Implications
• A herd PMWS diagnosis can be established when two criteria are fulfilled: an increase in postweaning mortality compared to the historical background in the herd, concurrent with observation of clinical signs compatible with PMWS; and diagnosis of PMWS in at least one of three to five pigs necropsied at that time.
• Diagnostic procedures must be implemented to exclude other potential causes of high mortality.
• SPC may be used to define a herd case diagnosis of PMWS when there is a significantly demonstrable increase in postweaning mortality compared to the historical background in that herd in association with clinical signs compatible with PMWS.
• SPC techniques require >20 data points to obtain reliable results.
Acknowledgements
This work was funded by Project No. 513928 from the Sixth Framework Programme of the European Commission. Authors are grateful to Miquel Collell for providing Spanish herd data.
References
1. Segalés J, Allan GM, Domingo M. Porcine circovirus diseases. Anim Health Res Rev. 2005;6:119–142.
2. Opriessnig T, Meng XJ, Halbur PG. Porcine circovirus type 2 associated disease: Update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Invest. 2007;19:591–615.
3. Harding J. Postweaning multisystemic wasting syndrome: Epidemiology and clinical presentation. J Swine Health Prod. 1998;6:249–254.
4. Harding JC. The clinical expression and emergence of porcine circovirus 2. Vet Microbiol. 2004;98:131–135.
5. Segalés J, Domingo M. Postweaning multisystemic wasting syndrome (PMWS) in pigs. A review. Vet Q. 2002;24:109–124.
6. Madec F, Rose N, Grasland B, Cariolet R, Jestin A. Post-weaning multisystemic wasting syndrome and other PCV2-related problems in pigs: a 12-year experience. Transboundary Emerg Dis. 2008;55:273–283.
7. Vigre H, Dohoo IR, Stryhn H, Jensen VF. Use of register data to assess the association between use of antimicrobials and outbreak of Postweaning Multisystemic Wasting Syndrome (PMWS) in Danish pig herds. Prev Vet Med. 2010;93:98–109.
8. Grau-Roma L, Fraile L, Segalés J. Recent advances in the epidemiology, diagnosis and control of diseases caused by porcine circovirus. Vet J. 2011;187:23–32.
9. Rosell C, Segalés J, Ramos-Vara JA, Folch JM, Rodríguez-Arrioja GM, Duran CO, Balasch M, Plana-Duran J, Domingo M. Identification of porcine circovirus in tissues of pigs with porcine dermatitis and nephropathy syndrome. Vet Rec. 2000;146:40–43.
10. McNeilly F, McNair I, O’Connor M, Brockbank S, Gilpin D, Lasagna C, Boriosi G, Meehan B, Ellis J, Krakowka S, Allan GM. Evaluation of a porcine circovirus type 2-specific antigen-capture enzyme-linked immunosorbent assay for the diagnosis of postweaning multisystemic wasting syndrome in pigs: comparison with virus isolation, immunohistochemistry, and the polymerase chain reaction. J Vet Diagn Invest. 2002;14:106–112.
11. Sorden S. Update on porcine circovirus and postweaning multisystemic wasting syndrome. Diagnostic notes. Swine Health Prod. 2000;8:133–136.
12. Grau-Roma L, Hjulsager CK, Sibila M, Kristensen CS, López-Soria S, Enøe C, Casal J, Bøtner A, Nofrarías M, Bille-Hansen V, Fraile L, Baekbo P, Segalés J, Larsen LE. Infection, excretion and seroconversion dynamics of porcine circovirus type 2 (PCV2) in pigs from post-weaning multisystemic wasting syndrome (PMWS) affected herds in Spain and Denmark. Vet Microbiol. 2009;135:272–282.
13. Fachinger V, Bischoff R, Jedidia SB, Saalmuller A, Elbers K. The effect of vaccination against porcine circovirus type 2 in pigs suffering from porcine respiratory disease complex. Vaccine. 2008;26:1488–1499.
14. Horlen KP, Dritz SS, Nietfeld JC, Henry SC, Hesse RA, Oberst R, Hays M, Anderson J, Rowland RR. A field evaluation of mortality rate and growth performance in pigs vaccinated against porcine circovirus type 2. JAVMA. 2008;232:906–912.
15. Pejsak Z, Pódgorska K, Truszczyński M, Karbowiak P, Stadejek T. Efficacy of different protocols of vaccination against porcine circovirus type 2 (PCV2) in a farm affected by postweaning multisystemic wasting syndrome (PMWS). Comp Immunol Microbiol Infect Dis. 2009;336:e1-e5.
16. Segalés J, Urniza A, Alegre A, Bru T, Crisci E, Nofrarías M, López-Soria S, Balasch M, Sibila M, Xu Z, Chu HJ, Fraile L, Plana-Duran J. A genetically engineered chimeric vaccine against porcine circovirus type 2 (PCV2) improves clinical, pathological and virological outcomes in postweaning multisystemic wasting syndrome affected farms. Vaccine. 2009;27:7313–7321.
17. Reneau JK, Lukas J. Using statistical process control methods to improve herd performance. Vet Clin North Am Food Anim Pract. 2006;22:171–193.
18. Fraile L, Crisci E, Weenberg J, Armadans M, Mendoza L, Ruiz L, Bernaus S, Montoya M. Effect of treatment with phytosterols in three herds with porcine respiratory disease complex. J Swine Health Prod. 2009;17:32–41.
19. Koketsu Y, Duangkaew C, Dial GD, Reeves DE. Within-farm variability in number of females mated per week during a one-year period and breeding herd productivity on swine farms. JAVMA. 1999;214:520–524.
20. Sarli G, Ostanello F, Morandi F, Fusaro L, Gnudi M, Bacci B, Nigrelli A, Alborali L, Dottori M, Vezzoli F, Barigazzi G, Fiorentini L, Sala V, Leotti G, Joisel F. Application of a protocol for the diagnosis of postweaning multisystemic wasting syndrome in Italy. Vet Rec. 2009;164:519–523.
21. Wallgren P, Hasslung F, Bergström G, Linder A, Belák K, Hård af Segerstad C, Stampe M, Molander B, Björnberg Kallay T, Nörregård E, Ehlorsson CJ, Törnquist M, Fossum C, Allan GM, Robertsson JÅ. Postweaning multisystemic wasting syndrome – PMWS. The first year with the disease in Sweden. Vet Q. 2004;26:170–187.
22. Gerry PQ, Michael J. Experimental Design and Data Analysis for Biologists. Cambridge, United Kingdom: Cambridge University Press. 2002;7–9.
23. Thrusfield M, Ortega C, de Blas I, Noordhuizen JP, Frankena K. WIN EPISCOPE 2.0: Improved epidemiological software for veterinary medicine. Vet Rec. 2001;148:567–572.
24. Mohammed MA, Worthington P, Woodall WH. Plotting basic control charts: tutorial notes for healthcare practitioners. Qual Saf Health Care. 2008;17:137–145. doi:10.1136/qshc.2004.012047.
25. Control of Porcine Circovirus Diseases (PCVDs): Towards improved food quality and safety. Sixth Framework Programme Priority SSP/5.4.6. Specific targeted research or innovation project. PMWS case definition (herd level). October 2005. Available at http://www.pcvd.eu/news.php. Accessed 6 February 2012.
26. Segalés J, Rosell C, Domingo M. Pathological findings associated with naturally acquired porcine circovirus type 2 associated disease. Vet Microbiol. 2004;98:137–149.
27. Walker RA. Quantification of immunohistochemistry – issues concerning methods, utility and semiquantitative assessment I. Histopathology. 2006;49:406–410.
*28. Jorsal SE, Bille-Hansen V, Vigre H, Larsen PB, Bøtner A, Nielsen EO, Enøe C, Bækbo P. PMWS– Laboratory diagnosis on herd and pig level in a Danish case-control study. Proc IPVS. Copenhagen, Denmark. 2006:270.
*29. Rademacher C. Use of statistical process control in finishing records. What’s Your Interpretation? J Swine Health Prod. 2004;12:158–159.
30. De Vries A, Reneau JK . Application of statistical process control charts to monitor changes in animal production systems. J Anim Sci. 2010;88:E11-E24. doi:10.2527/jas.2009-2622
* Non-refereed references.