Costa G, Oliveira S, Torrison J. Detection of Actinobacillus pleuropneumoniae in oral-fluid samples obtained from experimentally infected pigs. J Swine Health Prod. 2012;20(2):78–81
| Diagnostic notes | Peer reviewed |
SummaryThe use of cotton ropes has been recently proposed to collect oral fluids from pigs as a method to detect and monitor viral infections in swine populations. However, its use for detection and monitoring of swine bacterial pathogens by polymerase chain reaction (PCR) has not been assessed. In this study, oral-fluid testing for diagnosis of Actinobacillus pleuropneumoniae was evaluated over time, utilizing samples from experimentally infected pigs. Eighty pigs were randomly assigned to experimental groups infected with A pleuropneumoniae serovars 1, 3, 5, 7, 10, 12, or 15 and a non-inoculated control group. Oral fluids and blood samples were collected prior to infection, 1 day post infection, and weekly thereafter for 7 consecutive weeks. Oral fluids were tested for A pleuropneumoniae, Haemophilus parasuis, and Streptococcus suis by specific PCR tests offered by the Minnesota Veterinary Diagnostic Laboratory. Actinobacillus pleuropneumoniae was detected on days 1 and 7 post infection, whereas H parasuis and S suis, normal colonizers of the swine respiratory tract, were detected at all time points. These results indicate that oral-fluid testing has the potential to be a screening tool for detection of swine bacterial pathogens. Field studies are indicated to explore this potential further. | ResumenRecientemente, el uso de cuerdas de algodón se ha propuesto para colectar fluidos orales de cerdos como un método para detectar y monitorear infecciones virales en poblaciones porcinas. Sin embargo, su uso para la detección y monitoreo de patógenos bacterianos porcinos mediante la reacción en cadena de la polimerasa (PCR por sus siglas en inglés) no ha sido estudiada. En este estudio, se evaluaron a través del tiempo, los fluidos orales para el diagnóstico de Actinobacillus pleuropneumoniae, utilizando muestras de cerdos experimentalmente infectados. Ochenta cerdos fueron asignados al azar a grupos experimentales infectados con A pleuropneumoniae serovariedades 1, 3, 5, 7, 10, 12, ó 15 y a un grupo control no inoculado. Se colectaron fluidos orales y muestras de sangre antes de la infección, el día 1 post infección, y a partir de entonces, cada semana durante 7 semanas consecutivas. Se probaron los fluidos orales contra A pleuropneumoniae, Haemophilus parasuis, y Streptococcus suis con la prueba de PCR en el laboratorio de diagnóstico veterinario de Minnesota. El A pleuropneumoniae fue detectado los días 1 y 7 post infección, mientras que el H parasuis y S suis, colonizadores normales del tracto respiratorio porcino, fueron detectados en todos los muestreos. Estos resultados indican que la prueba de fluidos orales tiene el potencial de ser una prueba de discriminación para la detección de patógenos bacterianos porcinos. Se recomiendan estudios de campo para explorar este potencial con más profundidad. | ResuméL’utilisation de cordes de coton pour récolter du fluide oral provenant de porcs a récemment été proposée comme méthode pour détecter et surveiller les infections virales dans les populations porcines. Toutefois, son utilisation pour la détection et la surveillance des bactéries pathogènes porcines par réaction d’amplification en chaîne par la polymérase (PCR) n’a pas été évaluée. Dans la présente étude, l’utilisation de fluide oral pour le diagnostic d’Actinobacillus pleuropneumoniae a été évaluée dans le temps, à l’aide d’échantillons provenant de porcs infectés expérimentalement. Quatre-vingts porcs ont été répartis au hasard à des groupes expérimentaux infectés avec A pleuropneumoniae sérovars 1, 3, 5, 7, 10, 12, ou 15 et à un groupe témoin non-infecté. Des échantillons de fluide oral et de sang ont été prélevés avant l’infection, 1 jour post infection et à chaque semaine par la suite pour 7 semaines consécutives. Les échantillons de fluide oral ont été testés par PCR pour A pleuropneumoniae, Haemophilus parasuis, et Streptococcus suis au Minnesota Veterinary Diagnostic Laboratory. Actinobacillus pleuropneumoniae a été détecté aux jours 1 et 7 post infection, alors que H parasuis et S suis, qui colonisent normalement le tractus respiratoire porcin, ont été détectés lors de tous les prélèvements. Ces résultats indiquent que les tests effectués à l’aide de fluide oral ont le potentiel d’être utilisés comme outil de détection pour les bactéries pathogènes du porc. Des études en condition de terrain sont recommandées afin d’explorer ce potentiel. |
Keywords: swine, Actinobacillus pleuropneumoniae, diagnostic methods, oral fluids, polymerase chain reaction, APP, PCR
Search the AASV web site
for pages with similar keywords.
Received: June 16, 2011
Accepted: November 7, 2011
Actinobacillus pleuropneumoniae (APP) is a gram-negative late colonizer of the upper respiratory tract of swine. This agent is associated with development of swine pleuropneumonia, which is characterized by sudden death, severe lung necrosis and hemorrhage, and extensive fibrinous pleuritis.1 Infection is usually diagnosed by bacterial culture and serological testing.2 Unexpected serological results are further investigated by detection of APP in the tonsils by polymerase chain reaction (PCR).3 Collection of samples from live pigs, especially tonsillar swabs, may be labor intensive and requires animal restraint.
Recently, the use of cotton ropes has been proposed to collect oral fluids from pigs as an alternative method to detect and monitor viral infections.4,5 This method has been successful in detecting infections with porcine reproductive and respiratory syndrome virus, porcine circovirus type 2, and swine influenza virus under both experimental and field conditions.4,5 Oral-fluid testing has several advantages over traditional sampling. Besides being a non-invasive method, it reduces time, labor, and costs involved in sample collection and does not require intensive training of the farm staff.6 Oral-fluid testing has also been used to detect specific antibodies.7-9 However, its use for detection of clinically relevant swine bacterial pathogens has not been assessed.
In this study, we evaluated use of oral fluid as a sample for detection of A pleuropneumoniae in pigs experimentally infected with clinically relevant serovars. As a control, we compared detection of A pleuropneumoniae with that of Haemophilus parasuis and Streptococcus suis, which are normal colonizers of the upper respiratory tract and tonsils of healthy pigs. This was a parallel project conducted concurrently with a major experiment designed to evaluate diagnostic tools commercially available for detection of A pleuropneumoniae.10
Materials and methods
The protocol for animal use was approved by the University of Minnesota Institutional Animal Care and Use Committee.
This experiment was part of a major project10 in which 80 six-week-old pigs were randomly divided into eight groups of 10 pigs each. Pigs from Groups 1 to 7 were inoculated intranasally (1 mL per nostril) with 1 × 106 colony-forming units per mL of the following A pleuropneumoniae serovars: 1, 3, 5, 7, 10, 12, or 15. Pigs in Group 8 were not inoculated and remained as negative controls. All pigs were obtained from a source free of A pleuropneumoniae, determined by clinical history, tonsil PCR,11 and serological testing.2 Study groups were housed in separate rooms at the isolation facility of the University of Minnesota for 8 consecutive weeks, which included 1 week for acclimatization and 7 weeks for sample collection.
The reference strains used to experimentally infect naive pigs with A pleuropneumoniae serovars 1 (4074), 3 (1421), 5 (K17), and 7 (WF83) were obtained from the American Type Culture Collection. Reference strains for serovars 10 (22009), 12 (1096), and 15 (HS143) were kindly provided by Dr Pat Blackall.12 Six serovars were selected on the basis of their prevalence in US swine herds using 2002-2008 data from the Minnesota Veterinary Diagnostic Laboratory (MVDL), and serovar 10 was included due to frequent detection of antibodies against this serovar.
Unbleached cotton ropes (0.6 cm diameter) were purchased at a local hardware store. One-meter rope segments were hung in all eight isolation rooms at pig shoulder height for easy access. Ropes were placed in a clean area of the pen and away from water and feed. Pigs were allowed to chew on the ropes for 20 to 30 minutes just prior to inoculation (Day 0), Day 1, and weekly thereafter (Days 7, 14, 28, 35, 42, and 49).
Blood samples were collected on Day 0 and weekly thereafter. Sera were tested for A pleuropneumoniae antibodies using the Multi-APP test offered by the University of Montreal (Saint-Hyacinthe, Québec, Canada), which contains a mix of lipopolysaccharides of all APP serovars currently described. Results are reported as positive or negative.
All pigs were euthanized 49 days post infection, and postmortem examinations were conducted to evaluate the presence and severity of A pleuropneumoniae lesions.
Ropes containing oral fluids were placed individually into clean plastic bags and processed as previously described.5 Briefly, oral fluids were extracted by compressing the wet end of the rope in the plastic bag. A bottom corner of the bag was then clipped to drain the fluids into a 50-mL centrifuge tube. Collected oral-fluid samples were stored at 4°C until DNA extraction and bacterial isolation. Detection of H parasuis and S suis in oral fluids collected from all groups at all time points was used as a control for sample processing, DNA extraction, and potential presence of PCR inhibitors in oral fluids.
Deoxyribonucleic acid was extracted within 48 hours of oral-fluid collection. Aliquots of 1.0 mL obtained from each sample were placed into microcentrifuge tubes and vortexed for 15 seconds. The tubes were centrifuged at 28,350g for 3 minutes. Following centrifugation, the supernatant was discarded and cell pellets were resuspended in 200 μL of PrepMan reagent (Applied BioSystems, Foster City, California) and vortexed for 15 seconds. The suspensions were then boiled for 20 minutes and recentrifuged at 28,350g for 3 minutes. Fifty microliters of the supernatant from each tube was transferred to a new tube containing 50 μL of nuclease-free water. Extracted DNA was stored at 4°C overnight before being used as a template for PCR detection.
For A pleuropneumoniae isolation, a loopful of each oral-fluid sample was plated onto 5% sheep blood agar plates with a nurse Staphylococcus epidermidis streak. All plates were incubated at 37°C in a 5% CO2 atmosphere for 48 hours. Plates were examined for A pleuropneumoniae suspect colonies at 24 and 48 hours. Suspect colonies (hemolytic with satellitism to the S epidermidis nurse streak) were re-plated onto blood agar, also with a nurse S epidermidis streak, and incubated for 24 additional hours to confirm the identity of the isolate. Deoxyribonucleic acid from oral fluids and from suspected isolated colonies was tested by specific gel-based PCR tests routinely used by the MVDL for A pleuropneumoniae, H parasuis, and S suis. Detection of H parasuis and S suis was used as a “positive” control, as these two microorganisms are considered normal inhabitants of the pig’s upper respiratory flora.
Results
An average of 5 mL of oral fluids was recovered from each rope at each collection. Detection of A pleuropneumoniae, H parasuis, and S suis in oral fluids collected at different time points is shown in Figure 1. Actinobacillus pleuropneumoniae was detected by PCR in samples collected at Day 1 in the group inoculated with serovar 3, and at Day 7 in the groups inoculated with serovars 7 and 10. Actinobacillus pleuropneumoniae was not detected by PCR at any time point in samples from pigs inoculated with serovars 1, 5, 12, or 15. Actinobacillus pleuropneumoniae was not cultured from oral-fluid samples collected throughout the experiment. Detection of APP in oral-fluid samples by PCR and number of seropositive pigs in each group by Multi-APP ELISA are shown in Table 1.
Figure 1: Detection of Haemophilus parasuis, Streptococcus suis, and Actinobacillus pleuropneumoniae (APP) by polymerase chain reaction (PCR) in oral-fluid samples (one sample per room) collected at Days 0, 1, 7, 14, 28, 35, 42, and 49 post inoculation with APP. A total of 80 six-week-old pigs were randomly divided into eight groups of 10 pigs each, housed in different rooms and infected with seven different APP serovars (one serovar per group and a non-infected control group). APP was detected in oral-fluid samples at 1 day post inoculation in rooms housing pigs inoculated with serovar 3 and at 7 days post inoculation in rooms housing pigs inoculated with serovars 7 and 10.
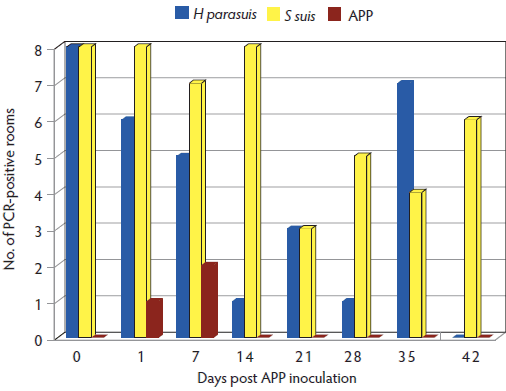 |
Table 1: Detection of APP in oral-fluid samples by PCR and number of seropositive pigs in groups of 10 pigs*
* In each group, at approximately 7 weeks of age, all 10 pigs were either inoculated with one APP serovar or were not inoculated (Control). Oral-fluid samples (one sample per group) were collected on Day 0 (before inoculation), on Day 1, and weekly thereafter. Blood samples were collected from each pig before inoculation on Day 0 and weekly thereafter. Blood samples were not collected from any group on Day 1, nor from groups infected with serovars 1 and 7 on Day 28. † Multi-APP ELISA; University of Montreal, Saint-Hyacinthe, Québec. ‡ Focal fibrous adhesions between the lung surface and the parietal pleura. § Hemorrhage in the dorsal lobes of the lungs (lesion characteristic of APP). APP = Actinobacillus pleuropneumoniae; PCR = polymerase chain reaction; ELISA = enzyme-linked immunosorbent assay; Neg = oral-fluid sample for the room was negative by PCR for APP; Pos = oral-fluid sample for the room was positive by PCR for APP; NA = data not |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Pigs inoculated with serovars 5 (n = 1), 7 (n = 4), 10 (n = 4), and 15 (n = 2) showed mild focal fibrous adhesions between the surface of the lungs and the parietal pleura at necropsy performed 49 days post inoculation. Pigs inoculated with serovars 7 and 10 also showed lung lesions (Table 1). No APP-specific lesions were observed in pigs inoculated with serovars 1, 3, or 12.
Discussion
Actinobacillus pleuropneumoniae infection was confirmed in pigs inoculated with serovars 1, 3, 7, 10, and 15. Pigs inoculated with serovars 5 and 12 and pigs from the control group were consistently negative by PCR and seronegative, which indicates that infection with these serovars was unsuccessful. Experimental infection of naive pigs with serovars 5 and 12 has been previously reported; however, higher challenge doses were used than in this study.13,14
Detection of H parasuis and S suis in oral fluids collected from all groups at all time points was used in this study as a control for sample processing, DNA extraction, and potential presence of PCR inhibitors in oral fluids. Detection of these two early colonizers at most time points was expected, as both are normal inhabitants of the upper respiratory tract of pigs.15,16 However, the differences in detection of H parasuis, S suis, and A pleuropneumoniae in oral fluids was an unexpected finding, especially because pigs were intentionally infected with APP. As tonsils are the main colonization site for this pathogen,17 APP was likely to be detected in oral fluids. However, some reports suggest that A pleuropneumoniae is primarily located deep in the tonsillar crypts, with lower numbers exposed on the surface of the tonsils,18 which may explain lack of detection in the oral-fluid samples and tonsil swabs.
Although H parasuis and S suis were consistently detected in oral fluids at different time points, a large variability was observed in the number of rooms positive for these bacteria throughout the study. Streptococcus suis was detected 49 times, whereas H parasuis was detected 31 times and APP was detected only three times. These data suggest that colonization dynamics and shedding of these microorganisms vary, and that shedding behavior differs considerably between early colonizers (H parasuis and S suis) and late colonizers (APP). Our results clearly show that there is a major difference in detection patterns among organisms and confirms that pathogen detection in oral fluids should be validated before being used for different pathogens.
Oral-fluid testing detected early infection in groups infected with A pleuropneumoniae serovars 3, 7, or 10, and this technique holds promise as a tool for broad screening of large populations. Oral-fluid testing detected infection earlier than serological testing in groups infected with serovars 3 and 7, so it could potentially be used as an additional tool to test pigs in isolation prior to introduction into naive populations. However, the apparent low sensitivity of oral-fluid testing for detecting APP is a limitation, and this method needs to be validated in field applications to determine whether it will be a practical diagnostic technique.
Implications
• Oral-fluid testing may be a valuable tool to screen swine populations not only for viruses, but also for bacterial pathogens.
• The use of oral-fluid testing is likely to play an important role in disease surveillance and monitoring of control and eradication strategies; however, this technique should be fully validated for each pathogen of interest, as the sensitivity of oral-fluid testing may vary among pathogens.
Acknowledgements
This work was supported in part by a grant from the National Pork Board. We would like to thank Dr Montserrat Torremorell and Dr Sagar Goyal for guidance and review of this paper.
References
1. Marsteller TA, Fenwick B. Actinobacillus pleuropneumoniae disease and serology. Swine Health Prod. 1999;7:161–165.
2. Dreyfus A, Schaller A, Nivollet S, Segers RP, Kobisch M, Mieli L, Soerensen V, Hussy D, Miserez R, Zimmermann W, Inderbitzin F, Frey J. Use of recombinant ApxIV in serodiagnosis of Actinobacillus pleuropneumoniae infections, development and prevalidation of the ApxIV ELISA. Vet Microbiol. 2004;99:227–238.
*3. Oliveira S, Rossow K, Olsen K, Collins J. Actinobacillus sp. biochemically and phenotypically similar to Actinobacillus pleuropneumoniae can be differentiated by genomic fingerprinting, toxin profiling, and sequencing of the 16S rRNA gene. Proc Am Assoc Vet Lab Diagnosticians. Reno, Nevada. 2007;136.
4. Prickett JR, Simer R, Christopher-Hennings J, Yoon KJ, Evans RB, Zimmerman JJ. Detection of porcine reproductive and respiratory syndrome virus infection in porcine oral fluid samples: a longitudinal study under experimental conditions. J Vet Diagn Invest. 2008;20:156–163.
5. Prickett JR, Kim W, Simer R, Yoon K-J, Zimmerman JJ. Oral-fluid samples for surveillance of commercial growing pigs for porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 infections. J Swine Health Prod. 2008;16:86–91.
6. Prickett JR, Zimmerman JJ. The development of oral fluid-based diagnostics and applications in veterinary medicine. Anim Health Res Rev. 2010;11:207–216.
7. Isaki L, Bairey M, van Patten L. Response of vaccinated swine to group E Streptococcus exposure. Cornell Vet. 1973;63:579–588.
8. Loftager MK, Eriksen L, Nielsen R. Antibodies against Actinobacillus pleuropneumoniae serotype 2 in mucosal secretions and sera of infected pigs as demonstrated by an enzyme-linked immunosorbent assay. Res Vet Sci. 1993;54:57–62.
9. Hyland K, Foss DL, Johnson CR, Murtaugh MP. Oral immunization induces local and distant mucosal immunity in swine. Vet Immunol Immunopathol. 2004;102:329–338.
10. Costa G, Oliveira S, Torrison J, Dee S. Evaluation of Actinobacillus pleuropneumoniae diagnostic tests using samples derived from experimentally infected pigs. Vet Microbiol. 2010;148:246–251.
11. Schaller A, Djordjevic SP, Eamens GJ, Forbes WA, Kuhn R, Kuhnert P, Gottschalk M, Nicolet J, Frey J. Identification and detection of Actinobacillus pleuropneumoniae by PCR based on the gene apxIVA. Vet Microbiol. 2001;79:47–62.
12. Blackall PJ, Klaasen HL, van den Bosch H, Kuhnert P, Frey J. Proposal of a new serovar of Actinobacillus pleuropneumoniae: serovar 15. Vet Microbiol. 2002;84:47–52.
13. Ueda Y, Ohtsuki S, Narukawa N. Efficacy of flor-fenicol on experimental Actinobacillus pleuropneumoniae in pigs. J Vet Med Sci. 1995;57:261–265.
14. Grondahl-Hansen J, Barfod K, Klausen J, Andresen LO, Heegaard PM, Sorensen V. Development and evaluation of a mixed long-chain lipopolysaccharide based ELISA for serological surveillance of infection with Actinobacillus pleuropneumoniae serovars 2, 6 and 12 in pig herds. Vet Microbiol. 2003;96:41–51.
15. Smart NL, Miniats OP, Maclnnes JI. Analysis of Haemophilus parasuis isolates from southern Ontario swine by restriction endonuclease fingerprinting. Can J Vet Res. 1988;52:319–324.
16. Amass SF, Clark LK, Wu CC. Source and timing of Streptococcus suis infection in neonatal pigs: Implications for early weaning procedures. Swine Health Prod. 1995;3:189–193.
17. Sidibé M, Messier S, Lariviere S, Gottschalk M, Mittal KR. Detection of Actinobacillus pleuropneumoniae in the porcine upper respiratory tract as a complement to serological tests. Can J Vet Res.1993;57:204–208.
18. Chiers K, Van Overbeke I, Donné E, Baele M, Ducatelle R, De Baere T, Haesebrouck F. Detection of Actinobacillus pleuropneumoniae in cultures from nasal and tonsillar swabs of pigs by a PCR assay based on the nucleotide sequence of a dsbE-like gene. Vet Microbiol. 2001;83:147–159.
* Non-refereed reference.