Penmetchsa TV, White BA, Maddox CW, et al. Molecular epidemiologic investigation of the role of gilts in the introduction and transmission of Salmonella in swine production systems. J Swine Health Prod. 2009;17(2):81–89
| Original research | Peer reviewed |
Cite as: Penmetchsa TV, White BA, Maddox CW, et al. Molecular epidemiologic investigation of the role of gilts in the introduction and transmission of Salmonella in swine production systems. J Swine Health Prod. 2009;17(2):81–89.
Also available as a PDF.
SummaryObjective: To investigate whether purchased gilts introduce new Salmonella genotypes into a swine production system and acquire Salmonella infection from resident pigs. Materials and methods: One multi-site and one single-site swine production system from Illinois were recruited on prior evidence of a high prevalence of Salmonella infection. Cohorts of 102 and 120 incoming gilts on the multi-site and single-site farms, respectively, were sampled longitudinally for five to eight visits until the gilts were introduced into the breeding herd. On each visit, fecal and floor samples were obtained from cohort gilts and an approximately equal number of resident pigs in the same or adjacent room or building. All samples were cultured to detect Salmonella. The 123 Salmonella isolates were genotyped using repetitive sequence polymerase chain reaction with REP, BOX, and ERIC primers. Cluster analysis classified the isolates by degree of genetic relatedness. The 45 isolates from the multi-site farm were serotyped. Results: For both farms, the most closely genetically related Salmonella isolates were obtained from the same room on the same visit. For the multi-site farm, there was no evidence of transmission between gilts and resident pigs. For the single-site farm, there was evidence of transmission between gilts and resident pigs in the breeding barns. However, conclusive evidence that incoming gilts introduce new Salmonella genotypes into a swine production system was lacking. Serotyping information supported conclusions drawn from genotyping data. Implications: Spatial separation of purchased gilts from other pigs by all-in, all-out pig flow reduces Salmonella transmission in swine production systems. | ResumenObjetivo: Investigar si las primerizas adquiridas introducen nuevos genotipos de Salmonella en un sistema de producción porcina y si contraen la infección de Salmonella de los cerdos residentes. Materiales y métodos: Un sistema de producción porcina de de sitios múltiples y uno de sitio único en Illinois se reclutaron ante la evidencia previa de una alta prevalencia de infección de Salmonella. Se tomaron muestras de manera longitudinal de grupos de 102 y 120 primerizas respectivamente, que entraron a un sistema multisitios y a uno de sitio único durante cinco a ocho visitas hasta que las primerizas se introdujeron al hato reproductor. En cada visita, se obtuvieron muestras fecales de primerizas y del piso del grupo en estudio y un número aproximadamente igual de cerdos residentes en el mismo cuarto o en el cuarto o edificio adyacente. Todas las muestras se cultivaron para detectar Salmonella. Los 123 aislados de Salmonella se genotipificaron utilizando la reacción en cadena de la polimerasa de secuencia repetitiva con primers de REP, BOX, y ERIC. El análisis de grupos clasificó a los aislados por su grado de relación genética. También se serotipificaron los 45 aislados de la granja de sitios múltiples. Resultados: En ambas granjas, los aislados de Salmonella genéticamente más relacionados se obtuvieron del mismo cuarto durante la misma visita. En la granja de sitios múltiples, no hubo evidencia de transmisión entre las primerizas y los cerdos residentes. En la granja de sitio único, hubo evidencia de transmisión entre las primerizas y los cerdos residentes en los edificios de reproducción. Sin embargo, faltó evidencia concluyente de que las primerizas adquiridas introducen genotipos de Salmonella al sistema de producción porcina. La información de serotipificación apoyó las conclusiones obtenidas del análisis de genotipificación. Implicaciones: La separación espacial de primerizas adquiridas de los cerdos mediante un flujo todo dentro-todo fuera reduce la transmisión de Salmonella en sistemas de producción porcina. | ResuméObjectif: Déterminer si les cochettes achetées introduisent de nouveau génotypes de Salmonella dans une structure de production porcine et si elles contractent une infection à Salmonella à partir des porcs résidents. Matériels et méthodes: Une structure de production multi-sites et une structure de production à site unique situées en Illinois, ont été sélectionnées compte tenu d’évidences d’une prévalence élevée d’infections par Salmonella. Des cohortes de 102 et 120 cochettes entrant, respectivement, sur la ferme à sites multiples et la ferme à site unique, ont été échantillonnées lors de cinq à huit visites jusqu’à ce que les cochettes soient introduites dans le troupeau reproducteur. À chaque visite, des échantillons fécaux et provenant du plancher ont été prélevés des cohortes de cochettes ainsi que d’un nombre approximativement égal de porcs résidents dans les mêmes chambres ou dans des chambres ou bâtisses adjacentes. Tous les échantillons ont été cultivés pour détecter la présence de Salmonella. Les 123 isolats de Salmonella obtenus ont été génotypés par réaction d’amplification en chaîne par la polymérase utilisant des séquences répétitives avec des amorces REP, BOX, et ERIC. L’analyse par agrégation a permis de classer les isolats en fonction de leur parenté génétique. Les 45 isolats provenant de la ferme à sites multiples ont été sérotypés. Résultats: Pour les deux fermes, les isolats de Salmonella les plus apparentés génétiquement provenaient de la même chambre au cours de la même visite. Pour la ferme multi-sites, il n’y avait aucune évidence de transmission entre les cochettes et les porcs résidents. Pour la ferme à site unique, il y avait évidence de transmission entre les cochettes et les porcs résidents dans les bâtiments de reproduction. Toutefois, il n’y avait pas d‘évidences claires que les nouvelles cochettes ont introduit de nouveaux génotypes de Salmonella dans les structures de production. Les résultats du sérotypage corroboraient ceux obtenus par génotypage. Implications: Une ségrégation spatiale entre des cochettes achetées et les autres porcs par une pratique de tout-plein, tout-vide réduit la transmission de Salmonella dans les structures de production porcine. |
Keywords: swine, gilt, Salmonella, transmission, molecular epidemiology
Search the AASV web site
for pages with similar keywords.
Received: February 15, 2008
Accepted: July 25, 2008
Salmonellosis in humans is a serious food-borne disease characterized by acute and chronic gastroenteritis, septicemia, and sometimes death. The United States Department of Agriculture (USDA) has placed Salmonella at the top of its priority list for elimination of food-borne pathogens, with pork identified as an important source of infection.1 Molecular biological analysis comparing the genotypes of human and pig isolates of Salmonella has repeatedly implicated pigs as a source of human infection.2-8
There have been numerous epidemiologic investigations of Salmonella in swine production systems. Salmonella has been isolated from samples of swine feces,9-15 pen floor contents,12-17 feed,12,13,15,16,18 water,14-16 boot swabs,14-16 flies,14-16 intestinal contents of rodents,14-16 and feces of birds14,15 and cats.14,15 Although these studies have identified numerous sources of Salmonella within the environment of the swine-production ecosystem, most of the focus on external sources for introduction of Salmonella into a production system has been on the role of feed, with the implication that this is the main conduit for introduction of Salmonella into a production system.10,18,19 However, several epidemiologic studies have found little or no Salmonella in feed.9,13,14 Thus, other sources for the introduction of Salmonella into a production system need to be investigated.
Purchase of gilts may be an important mechanism for introducing Salmonella into a herd without apparent clinical signs. Modern swine production systems typically replace 40% to 50% of sows each year.20,21 Monitoring suppliers of replacement breeding stock is a key component of national Salmonella control programs for swine production in Sweden and Denmark.20
In a prior study of the role of gilts in Salmonella transmission in swine production systems, the movement of two cohorts of 6-month-old gilts (n = 202) from a giltdevelopment farm to a breeding farm was monitored.22 Fecal samples were collected from gilts within 1 week before leaving the gilt-development farm and again within 2 weeks after arrival at the breeding farm. The rate of fecal shedding of Salmonella by the gilts was higher after transport and introduction to the breeding herd (> 20% after versus < 5% before). Serotyping of Salmonella isolates indicated that 55% of isolates from gilts at the breeding farm were serotypes not identified from gilt samples on the development farm. It was noted that new serotypes may have been identified only at the breeding farm because of increased shedding of Salmonella by gilts after transport, but that this may have also been due to acquisition of new infections from the environment of the breeding farm. Regardless of the source of infection, transportation of gilts was implicated as responsible for increasing the Salmonella load in the environment and thus maintaining a high prevalence of Salmonella infection in the herd. Transportation of pigs has been recognized repeatedly as a factor increasing the rate of Salmonella shedding,23-25 so the acquisition of gilts from an external source is likely to be a risk factor for introduction of new Salmonella into a herd.
The previous study by Davies et al22 suggests that introduction of 6-month-old gilts may contribute to the introduction and maintenance of Salmonella infections in swine herds. This increase in prevalence might be attributed to incoming gilts acquiring infection from resident pigs or vice versa. However, the role of gilts in transmitting Salmonella infections to herd residents was not investigated because resident pigs were not sampled. Also, a common practice in gilt acquisition is to purchase replacement breeding stock at weaning.26 Several studies have indicated that weaning-age pigs are a relatively minor source of Salmonella infection,11,12,27,28 most likely due to maternally acquired immunity,29 which provides justification for acquiring replacement gilts at weaning.
The present study was designed to extend the analysis of the role of acquired gilts in introducing Salmonella into a herd by also examining Salmonella shedding in resident pigs. Evaluation of transmission between incoming gilts and resident pigs was made more precise by using molecular genotyping30 and statistical methods developed for inferring transmission from genetic relatedness.15
Materials and methods
The project protocol was approved for animal use by the University of Illinois Institutional Animal Care and Use Committee, which was accepted by USDA for their purposes. One multi-site (Farm M) and one single-site (Farm S) commercial swine production facility in Illinois were investigated. Both farms had total confinement housing with concrete floors in the growing-finishing and breeding areas and slotted wood floors in the nursery. The farms were selected on the basis of prior evidence of a high prevalence of Salmonella. Farm M had the highest prevalence of Salmonella infection in a previous study of 18 swine farms in Illinois.15 Farm S had been identified through the Illinois Department of Agriculture because of outbreaks of enteric disease in the nursery due to Salmonella enterica within the previous year. Both farms acquired gilts at 2 to 3 weeks of age at intervals of ≤ 3 months, with at least 100 gilts per shipment. No Salmonella vaccine was used on either farm during the course of the study. On both farms, nursery-stage pigs received antibiotics in feed to promote growth.
Farm M had 3500 sows. Incoming gilts were isolated in a nursery on a separate site reserved for purchased gilts. At approximately 9 to 10 weeks of age, these gilts were moved as a cohort to a vacant room on one of several available finishing sites, where they remained until about 5 to 6 months of age. They were then moved to a gestation barn on one of two breeding sites, where cohort gilts were mixed in pens with “resident sows” already circulating in the breeding herd.
Farm S had 6000 sows. A cohort of incoming gilts from a single source was segregated from other pigs in one of two nursery buildings containing only one room. At 6 to 8 weeks of age, the cohort of gilts was moved to a single pen in a growing facility consisting of one large room with multiple pens housing pigs from various sources segregated by age. Pigs were moved as a cohort from pen to pen as they advanced in age, and moved to a similar finishing building at 14 to 16 weeks of age. At 5 to 6 months, each finishing-age gilt showing signs of estrus was moved into a pen shared with approximately 15 to 25 other gilts, in one of two breeding barns.
Sample collection
The first visit to a farm was on the day of gilt arrival. A cohort of incoming gilts was selected and ear-tagged for follow-up. Cohort size was 102 on Farm M and 120 on Farm S. On each visit, samples were collected from cohort pigs and from an approximately equal number of resident pigs that were the nearest neighbors (same room, adjacent room, or adjacent building). Subsequent farm visits were made as indicated in Tables 1 and 2. Samples collected included feces and swabs of pen floors. Feces were obtained from nursery pigs using feline fecal loops. From older pigs, a 1-g fecal sample was obtained by digital insertion into the anus, with the collector wearing a latex glove that was changed between pigs. Pen floors were sampled to obtain at least 1 g of floor contents by swabbing with one 4 × 4-inch cotton gauze for each pen, collecting in several parts of the floor where feces was deposited. Fecal samples were transferred immediately into tubes containing 9 mL tetrathionate broth (TTB) with iodine, and floor samples were placed in tubes with 25 mL TTB with iodine for enrichment of Salmonella. Tetrathionate broth tubes were placed on ice packs in coolers and transported to the laboratory the same day.
Table 1: Characteristics of samples collected from cohorts of replacement gilts for isolation of Salmonella at each visit to Farm M, a 3500-sow, multi-site commercial swine-production facility in Illinois*
* Replacement gilts were acquired at 2 to 3 weeks of age at intervals of ≤ 3 months, with ≥ 100 gilts per cohort. Cohort pigs were ear-tagged on the day of arrival. On each visit, fecal samples were collected from all cohort pigs and from approximately equal numbers of the nearest resident pigs. |
Table 2: Characteristics of samples collected for Salmonella isolation at each visit to Farm S, a 6000-sow, single-site commercial swine-production facility in Illinois*
* Replacement gilts were acquired at 2 to 3 weeks of age at intervals of ≤ 3 months, with ≥ 120 gilts per cohort. Cohort pigs were ear-tagged on the day of arrival. On each visit, fecal samples were collected from all cohort pigs and from approximately equal numbers of the nearest resident pigs. |
Bacteriologic culture and polymerase chain reaction analysis
Upon return to the laboratory (maximum of 8 hours after collection), all samples were incubated at 37°C for 48 hours. Thereafter, 100 μL of TTB broth from each tube was transferred to 10 mL Rappaport’s medium (R-10) and incubated for 24 hours at 37°C. Samples from R-10 were streaked onto xylose-lysine-tergitol 4 agar (XLT-4) plates and incubated for 24 hours at 37°C. Red colonies with black centers were picked and streaked onto brilliant green agar (BGA) plates. Red non-lactose-fermenting suspected Salmonella colonies from BGA plates were transferred to tryptic soy agar (TSA) plates and analyzed using polymerase chain reaction (PCR) with primers for the invA gene,31 using methods described previously.32 DNA was extracted by boiling for 5 minutes the cells obtained from an isolated colony from a TSA plate. Polymerase chain reaction-positive isolates were frozen at -80°C in tryptic soy broth for storage. Salmonella isolates were genotyped using repetitive sequence PCR (rep-PCR) methods with three different primers (REP, BOX, and ERIC) as described previously.30 Resulting DNA amplicons were separated by size by gel electrophoresis.
All Salmonella-positive isolates from Farm M were submitted to the National Veterinary Diagnostic Laboratory (Ames, Iowa) for serotyping.
Data analysis
Data analysis consisted of pairwise comparison of sizes of PCR amplicons to calculate the percentage of fragments matching by size (Dice coefficient) as an estimate of genetic distances between isolates.30 A 5% tolerance level for matching was used. Hierarchical cluster analysis, using the complete linkage algorithm, was conducted to determine the overall pattern of genetic similarity among isolates.33 On the basis of statistical principles of identification of clusters from the distribution of linkage distances,33 previous studies investigating cluster formation from a matrix of genetic distances calculated from three-primer rep-PCR had identified isolates within a genetic distance ≤ 0.15 as “similar.”15,30 In the present study, isolates within this distance were considered to have a recent common ancestor, thus implicating transmission from one source to another. Linkage of genetically similar isolates was examined with respect to their similarity in time of sampling (farm visit), location (same room or not), and whether a sample was from a resident pig or incoming gilt, using the binomial test,34 in an analytic model developed previously.15 For Farm M, the associations of serotypes with time of sampling, farm site, and porcine source of sample were tested using 2 × 2 contingency tables, as described previously.15 Due to use of multiple statistical tests, an alpha level for significance equal to .01 was used.
Results
There was no evidence of clinical salmonellosis on either farm during the course of the study, ie, no widespread signs of enteric disease. On Farm M, 856 samples were collected and cultured for Salmonella, 45 (5.6%) of which were positive (Table 3). Salmonella-positive fecal samples were found only in 22-week-old pigs at the breeding site. None of the fecal samples collected from 2- to 3-week-old nursery pigs or the floor samples from their pens were positive. However, Salmonella was detected in the floor samples after pigs were moved to the growing-finishing sites at 9 weeks of age. Not differentiating by location, pig source, or time of visit, Salmonella prevalence was higher in floor samples (44.0%) than in fecal samples (2.0%).
Table 3: Salmonella prevalence in a total of 856 fecal and floor samples collected from Farm M*
* Farm M and sampling schedule described in Table 1. ND = not done; NA = not applicable. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Serotypes of Salmonella isolates were clearly differentiated by site on Farm M. Three serotypes were identified from samples collected on finishing site 2 during visits 3 and 4: Alachau (n = 12), Anatum (n = 8), and Minnesota (n = 1). Three different serotypes were identified from samples collected on breeding site 3 during visit 5: Derby (n = 20), Typhimurium (n = 3), and Infantis (n = 1). There was also some differentiation of Salmonella serotype by pig source, with Typhimurium found only in the feces of 22-week-old cohort gilts, and Alachau found only on the pen floors of 9- and 10-week-old resident pigs.
On Farm S, 2345 samples were collected and cultured for Salmonella, 78 (3.3%) of which were positive (Table 4). Positive samples were found on each visit. There was no apparent differentiation of positive samples by location, pig source, or time of visit. Overall, Salmonella prevalence was higher among floor samples (5.8%) than fecal samples (2.5%). Serotyping information was not available for Salmonella isolated on Farm S.
Table 4: Salmonella prevalence in a total of 2345 fecal and floor samples collected from Farm S*
* Farm S and sampling schedule described in Table 2. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
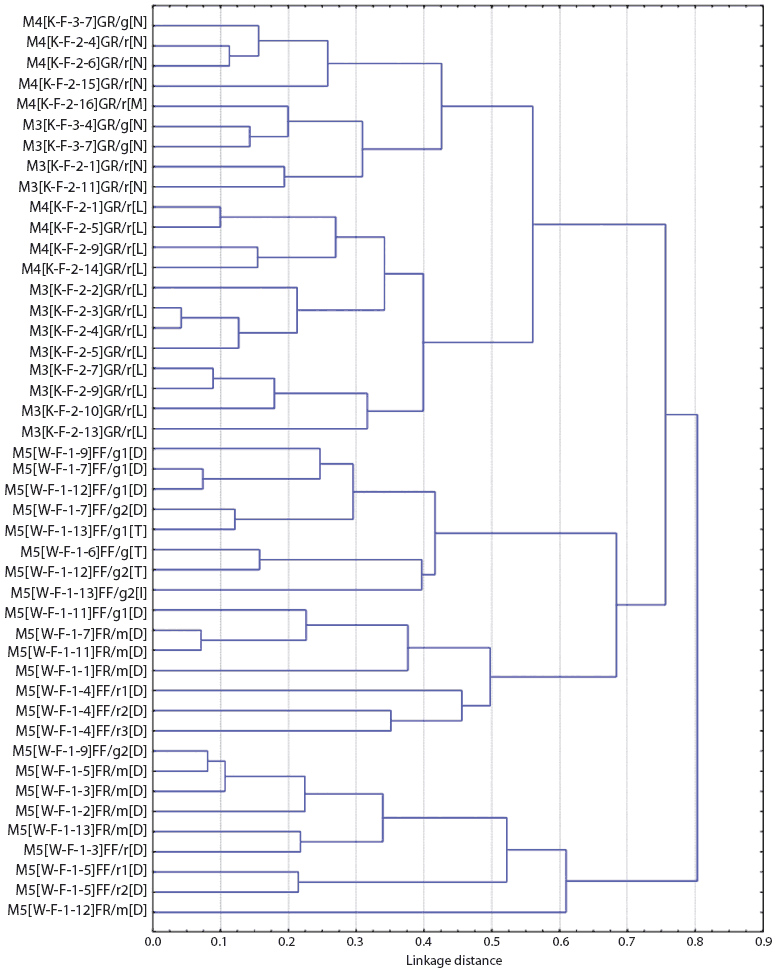
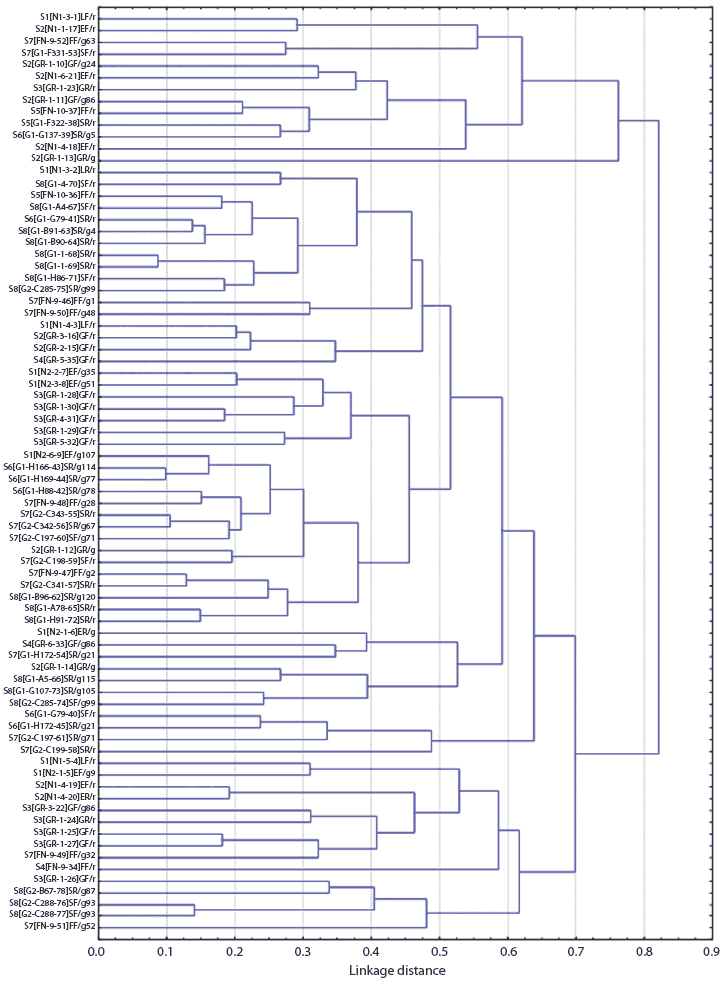
Figures 1 and 2 show the dendrograms from the cluster analysis of genetic distances between Salmonella isolates. For Farm M, of the three largest clusters formed (separated at a distance approximately equal to 0.7, or less than 30% similarity in rep-PCR fragmentation), two were from the breeding site (W) and one was from the finishing site (K). Within the large finishing-site cluster, there were two smaller clusters distinct at a distance approximately equal to 0.55, or about 45% rep-PCR similarity. One cluster included only isolates from the pen floors of resident pigs, consisting entirely of Salmonella isolates with the Alachau serotype. The other cluster included isolates from samples from the floors of both cohort gilts and resident pigs, all with the Anatum serotype, except for one isolate with the Minnesota serotype (Figure 1). Although this differentiation of serotypes by genetic cluster was clear, there were a few cases of isolates of different serotypes (Minnesota and Anatum; Derby and Typhimurim; Derby and Infantis) being more closely related to each other than to isolates of the same serotype. The genetic clusters of Salmonella isolates from Farm S were generally mixed with respect to farm visit and building (Figure 2).
| Figure 1: Dendrogram from cluster analysis of genetic
distances among Salmonella isolates from Farm M (described
in Table 1), as determined by repetitive sequence polymerase chain
reaction. Sample identification code: FV[S-B-R-P]ET/X#[Z], where F
= farm code, V = visit number, S = site (K = grower-finisher, W =
breeding), B = building code, R = room number, P = pen number, E =
stage of production (G = growing, F = finishing), T = sample class
(F = fecal, R = floor), X = pen type (g = cohort gilts only, r =
resident pigs only, m = mixed-cohort gilts and resident pigs), # =
isolate sequence number within pen for pens with more than one
isolate. [Z] indicates serotype code: D = Derby; I = Infantis, L =
Alachau, M = Minnesota, N = Anatum, T = Typhimurium.
|
| Figure 2: Dendrogram from cluster analysis of genetic
distances among Salmonella isolates from Farm S (described
in Table 2), as determined from repetitive sequence polymerase
chain reaction. Sample identification code: FV[B-P-#]ET/Xt, where F
= farm code, V = visit number, B = building code (N1, N2 =
nurseries; GR = growing stage, FN = finishing stage; G1, G2 =
gestation), P = pen number, # = sample sequence number, E = stage
of production (G = growing stage, F = finishing stage, S =
gestation), T = sample class (N = nursery pig, G = growing-stage
pig, F = finishing-stage pig, L = sow, R = floor), X = pen type (g
= pen with cohort gilts only, r = pen with resident pigs only, m =
mixed-cohort gilts and resident pigs), t = cohort gilt ear-tag
number.
|
Statistical analysis of the spatial, temporal, and pig characteristics of the most genetically similar isolates (genetic distance ≤ 0.15) revealed the following. For all 11 of the closest isolate pairs from Farm M, both isolates were collected from the same room on the same visit, and both isolates were from the same pig source, ie, either both from resident pigs or both from cohort gilts (P < .001, all cases). For Farm S, of the eight closest isolate pairs, six (75%) were identical for time of visit (P < .001), and six (75%) were identical for location (room) of sampling (P = .001). Three were different for pig source (ie, paired isolates were from both gilt and resident), but this association was not significant (P = .35). The mixed-source (gilt and resident) close isolate pairs were all sampled in the breeding barns.
For Farm M, where serotyping information was available, there were associations of the Derby serotype with the breeding site (P < .001) and the Alachau (P < .001) and Anatum (P < .001) serotypes with the finishing site. Also, samples from the pen floors of resident pigs were more likely to have Salmonella of the Alachau serotype than were samples from the pen floors of cohort gilts (P < .001).
Discussion
For both farms studied, the most genetically similar Salmonella isolates were found at the same farm visit and in the same housing unit. For the multi-site farm, Salmonella serotypes were differentiated by site. Isolates obtained on the same visit from different locations were genetically different, although not necessarily differentiated by serotype. The genotyping results are further confirmation of previous results,15 indicating the importance of physical barriers in inhibiting Salmonella transmission. One important difference between the two farms was that for the single-site farm only, there were several genetically similar isolate pairs from both gilt and resident pig samples, all from the gestation area. This indicates transmission of Salmonella between purchased gilts and resident pigs, although the direction of transmission is unknown. There was mixing of resident pigs and purchased gilts within the breeding barns on both farms. Transmission of Salmonella between gilts and residents on the single-site farm may have been due to earlier mixing within the same room in the grower stage. Only two farms were studied here, therefore limiting the ability to generalize. Nevertheless, the consistent finding, here and in the previous study,15 that physical barriers reduce the probability of transmission, suggests that the practice of mixing pigs from different sources in growing and finishing stages facilitates the transmission of Salmonella that is present within the swine production system. However, even when purchased gilts are moved as segregated cohorts through several housing facilities in the different stages of production (eg, nursery to grow-finish to gestation), the success of preventing Salmonella transmission between housing units depends upon additional factors, such as disinfection procedures and limiting the flow of air, water, human traffic, and animal vectors such as rodents, birds, cats, and insects.14
Serotyping information on Salmonella isolates from the multi-site farm was insufficient to provide accurate data on transmission. Three isolates obtained from cohort gilts on the finishing site shared the Anatum serotype with five isolates obtained from resident pigs on the same site; however, their genetic distances based on rep-PCR were < 60% similarity in DNA fragment matching, indicating they were unlikely to be related to a recent common source.
Definitive evidence for introduction of new genotypes of Salmonella into a swine production system through importation of gilts was not provided by the present study. The two farms studied provided contrasting evidence on the risk of purchased gilts being infected with Salmonella. Failure to detect shedding of Salmonella from nursery-stage pigs on the multi-site farm suggests that pigs of this age may be at low risk for contributing to the Salmonella load in the production system. In contrast, detection of Salmonella in fecal and floor samples from newly arrived gilts on the single-site farm identifies purchased gilts as a possible source of new Salmonella infection for a production system. Previous studies have reported low rates of Salmonella shedding in nursery-age pigs.13,29 These low shedding rates may be attributable either to maternally acquired immunity or to the smaller amounts of feces collected from 2- to 3-week-old piglets.35 Application of antibiotics to feed is also likely to inhibit Salmonella excretion. However, smaller pigs defecate less, and therefore shed less Salmonella into a production system environment, all other things being equal. Thus, recently weaned gilts with maternal antibodies, less history of exposure to pathogens, receiving oral antibiotics, and with lower fecal output, are less likely to introduce new Salmonella genotypes into a production system than are gilts acquired as replacements at 4 to 5 months of age. Purchase of early-weaned gilts has been recognized as a strategy for reducing Salmonella transmission.27
From 1996 to 2000, the replacement rate of breeding herd females in US commercial swine herds averaged near 50%; average herd parity of culled sows ranged from 3.1 to 3.7.21 High culling rates result in purchase of more breeding gilts. Swine represent the largest reservoir of Salmonella on any farm, and incoming gilts represent a large external biomass introduced into any swine-production ecosystem, therefore increasing the risk of bringing new infections onto the farm. Thus, gilt purchase becomes an important factor in making decisions about disease control.
In the previous study22 of the role of gilts in introducing Salmonella into a herd, cultures of fecal samples from recently transported 6-month-old gilts provided evidence for the appearance of serotypes of Salmonella not present on the source farm. From this it was inferred that transmission of Salmonella from resident pigs to gilts had occurred, although this was recognized as inconclusive because resident pigs had not been sampled.22 The higher Salmonella shedding rate (> 20%) for 6-month-old gilts in the previous study22 might be indicative of an overall higher risk of acquiring and shedding Salmonella in this age group than in the recently weaned pigs in the present study, in which Salmonella shedding was not observed. If evidence from future studies confirms these age differences in Salmonella shedding rate after transportation, the case for reducing Salmonella infection in a herd by acquisition of gilts at an early age will be strengthened.
Compared to the previous study22 of gilt introduction to the herd, in which pathways of transmission were inferred from the results of serotyping, an important improvement in the present study is the use of molecular genotyping to link Salmonella isolates of a common origin. Previous studies15,30,36 have reported that rep-PCR provides better discriminative ability among Salmonella isolates than serotyping, with the latter often failing to differentiate genetically distinct isolates. The comparison of rep-PCR and serotyping results from the multi-site farm in the present study provides additional evidence indicating that Salmonella serotyping lacks the precision necessary to definitively track Salmonella transmission in swine production systems.
In the study of Salmonella transmission in a complex ecosystem such as a swine production unit, only a small fraction of the Salmonella present can be detected because of practical sampling limitations. Even for infected pigs, due to intermittent shedding, standard sampling of feces and pen floor contents can only identify recent Salmonella excretion. Thus, infected pigs not excreting Salmonella at the time of sampling, but which may have been the source or destination for Salmonella transmission, may not be detected. Another limiting factor is the imperfect sensitivity of standard laboratory culturing methods for detection of Salmonella.37 Several PCR procedures for more direct detection of Salmonella in feces have been developed, but they have low sensitivity for field samples with low Salmonella loads, and thus an enrichment stage is necessary.37-39 Several studies have reported that, after enrichment, various PCR methods have high sensitivity and agree well with standard culture methods.40-42 However, an evaluation of field samples of bovine feces using the same invA PCR as that used in the present study, and culture methods similar to those used in the present study, found standard bacterial culture of Salmonella to have only 75% sensitivity compared to PCR on samples processed through enrichment media.39 These results indicate that reserving the invA PCR until Salmonella isolates have been obtained may result in falsely classifying some samples as negative. Complete culture, including plating on differential and selective media, is still required to recover Salmonella for genotyping, which is needed for the study of transmission. Use of PCR immediately after selective enrichment broth culture not only increases sensitivity in detecting Salmonella, but provides savings by not processing negative samples on differential and selective agar media.39 This savings is offset somewhat by use of PCR on the large number of negative samples expected in field investigations.
The analysis conducted here provides confirmatory evidence for transmission of Salmonella between pigs from different sources, although it was not possible to determine whether the Salmonella transmitted was from outside the production system or was already circulating within the environment. As this study included only two swine production systems, additional study is needed to determine the specific risk of introducing Salmonella when 2- to 3-week-old gilts are purchased.
Implications
- Transmission of Salmonella occurs primarily within the same housing unit; thus, spatial separation of purchased gilts from other pigs by all-in, all-out pig flow reduces Salmonella transmission in swine production systems.
- Although transportation stress induces Salmonella shedding, gilts purchased at weaning age are less likely to shed Salmonella than older gilts and thus are preferred for preventing introduction of Salmonella into a production system.
- Mixing pigs from different sources in gestation is likely to increase Salmonella prevalence and genetic diversity in the breeding herd and should be taken into account in the management of movement of sows through the breeding herd.
Acknowledgement
This work was supported by US Department of Agriculture project on “Molecular Epidemiology of Salmonella transmission in different swine production systems” (1999) and by the National Pork Board and the University of Illinois Research Board.
References
1. Davies PR. Food safety and its impact on domestic and export markets. Swine Health Prod. 1997;5:13–20.
2. Maguire HCF, Codd AA, Mackay VE, Rowe B, Mitchell E. A large outbreak of human salmonellosis traced to a local pig farm. Epidemiol Infect. 1993;110:239–246.
3. Nastasi A, Mammina C, Villafrate MR. Epidemiology of Salmonella typhimurium: ribosomal DNA analysis of strains from human and animal sources. Epidemiol Infect. 1993;110:553–565.
4. Baggesen DL, Wegener HC. Phage types of Salmonella enterica ssp. enterica serovar Typhimurium isolated from production animals and humans in Denmark. Acta Veterinaria Scand. 1994;35:349–354.
5. Wegener HC, Baggeson DL. Investigation of an outbreak of human salmonellosis caused by Salmonella enterica ssp. enterica serovar Infantis by use of pulsed field gel electrophoresis. Int J Food Microbiol. 1996;32:125–131.
6. Murase T, Okitsu R, Suzuki H, Morozumi H, Matsushima A, Nakamura A, Yamai S. Evaluation of DNA fingerprinting by PFGE as an epidemiologic tool for Salmonella infections. Microbiol Immunol. 1995;39:673–676.
7. Valdezate S, Vidal A, Herrera-Leon S, Pozo J, Rubio P, Usera MA, Carvajal A, Echeita MA. Salmonella Derby clonal spread from pork. Emerg Infect Dis. 2005;11:694–698.
8. Buchholz U, Brodhun B, Brockmann SO, Dreweck CM, Prager R, Tscäpe H, Ammon A. An outbreak of Salmonella München in Germany associated with raw pork meat. J Food Prot. 205;68:273–276.
9. Davies PR, Morrow WE, Jones FT, Deen J, Fedroka-Cray PJ, Harris IT. Prevalence of Salmonella in finishing swine raised in different production systems in North Carolina, USA. Epidemiol Infect. 1997;119:237–244.
10. Davies PR, Morrow WE, Jones FT, Deen J, Fedroka-Cray PJ, Gray JT. Risk of shedding Salmonella organisms by market-age hogs in a barn with open flush gutters. JAVMA. 1997;210:386–389.
11. Davies PR, Bovee FG, Funk JA, Morrow WE, Jones FT, Deen J. Isolation of Salmonella serotypes from feces of pigs raised in a multiple-site production system. JAVMA. 1998;212:1925–1929.
12. Davies PR, Funk JA, Morrow WEM. Fecal shedding of Salmonella by a cohort of finishing pigs in North Carolina. Swine Health Prod. 1999;7:231–234.
13. Funk JA, Davies PR, Nichols MA. Longitudinal study of Salmonella enterica in growing pigs reared in multiple-site swine production systems. Vet Microbiol. 2001;83,45–60.
14. Barber DA, Bahnson PB, Isaacson RE, Jones CJ, Weigel RM. Distribution of Salmonella in swine production ecosystems. J Food Prot. 2002;65:1861–1868.
15. Weigel RM, Nucera D, Qiao B, Teferedegne B, Suh DK, Barber DA, Bahnson PB, Isaacson RE, White BA. Testing an ecological model for transmission of Salmonella enterica in swine production ecosystems using genotyping data. Prev Vet Med. 2007;81:274–289.
16. Letellier A, Messier S, Paré J, Ménard J, Quessy S. Distribution of Salmonella in swine herds in Québec. Vet Microbiol. 1999:67:299–306.
17. Van de Wolf PJ, Lo Fo Wong DMA, Wolbers WB, Elbers ARW, van der Heijden HMJF, van Schie FW, Hunneman WA, Willeberg P, Tielen MJM. A longitudinal study of Salmonella enterica infections in high- and low-seroprevalence finishing swine herds in the Netherlands. Vet Quart. 2001;23:116–121.
18. Harris IT, Fedroka-Cray PJ, Gray JT, Thomas LA, Ferris K. Prevalence of Salmonella organisms in swine feed. JAVMA. 1997;210:382–385.
19. Fedorka-Cray PJ, Hogg A, Gray JT, Lorenzen K, Velasquez J, von Behren P. Feed and feed trucks as sources of Salmonella contamination in swine. Swine Health Prod 1997;5:189–193.
20. Mousing J, Jensen PT, Halgaard C, Bager F, Feld N, Nielsen B, Nielsen JP, Bech-Nielsen S. Nationwide Salmonella enterica surveillance and control in Danish slaughter swine herds. Prev Vet Med. 1997;29:247–261.
*21. Koketsu Y, ed. PigCHAMP DataSHARE, 2000 Summary Report. St Paul, Minnesota: University of Minnesota; 2000.
22. Davies PR, Funk JA, Morrow WEM. Fecal shedding of Salmonella by gilts before and after introduction to a swine breeding farm. Swine Health Prod. 2000;8:25–29.
23. Isaacson RE, Firkins L, Weigel RM, Zuckerman F, DiPietro J. Effect of transportation and feed withdrawal on the presence of Salmonella enterica in pigs. Am J Vet Res. 1999;60:1155–1158.
24. Larsen ST, McKean JD, Hurd HS, Rostagno MH, Griffith RW, Wesley IV. Impact of commercial preharvest transportation and holding on the prevalence of Salmonella enterica in cull sows. J Food Protect. 2003;66:1134–1138.
25. Gebreyes WA, Davies PR, Turkson P, Morrow WEM, Funk JA, Altier C. Salmonella enterica serovars from pigs on farms and after slaughter and validity of using bacteriologic data to define herd Salmonella status. J Food Prot. 2004;67:691–697.
26. Dee SA. An overview of production systems designed to prepare naïve replacement gilts for impending PRRSV challenge: A global perspective. Swine Health Prod. 1997;5:231–239.
27. Berends BR, Urlings HAP, Snijders JMA, van Knapen F. Identification and quantification of risk factors in animal management and transport regarding Salmonella spp. in pigs. Int J Food Microbiol. 1996;30;37–53.
28. Dahl J, Wingstrand A, Neilsen B, Baggesen DL. Elimination of Salmonella typhimurium infections by strategic movement of pigs. Vet Rec. 1997;140:679–681.
29. Kranker S, Alban L, Boes J, Dahl J. Longitudinal study of Salmonella enterica serotype Typhimurium infection in three Danish farrow-to-finish swine herds. J Clin Microbiol. 2003;41:2282–2288.
30. Weigel RM, Qiao B, Teferedegne B, Suh DK, Barber DA, Isaacson RE, White BA. Comparison of pulsed field gel electrophoresis and repetitive sequence polymerase chain reaction as genotyping methods for detection of genetic diversity and inferring transmission of Salmonella. Vet Microbiol. 2004;100:205–217.
31. Chiu CH, Ou JT. Rapid identification of Salmonella
serovars in feces by specific detection of virulence genes,
invA and spvC, by an enrichment broth
culture-multiplex PCR combination assay.
J Clin Microbiol. 1996;34:2619–2622.
32. Nucera DM, Maddox CW, Hoien-Dalen P, Weigel RM. Comparison of API 20E and invA PCR for identification of Salmonella enterica isolates from swine production units. J Clin Microbiol. 2006;44:3388–3390.
33. Anderberg MR. Cluster Analysis for Applications. New York: Academic Press; 1973.
34. Fleiss JL, Levin B, Paik MC. Statistical Methods for Rates and Proportions. 3rd ed. Hoboken, New Jersey: John Wiley & Sons; 2003.
35. Funk JA, Davies PR, Nichols MA. The effect of fecal sample weight on the detection of Salmonella enterica in swine feces. J Vet Diag Invest. 2000;12:412–418.
36. Johnson JR, Clabots C, Azar M, Boxrud DJ, Besser JM, Thurn JR. Molecular analysis of a hospital cafeteria-associated salmonellosis outbreak using modified repetitive element PCR fingerprinting. J Clin Microbiol. 2001;39:3452–3460.
37. Malorny B, Hoorfar J. Toward standardization of diagnostic PCR testing of fecal samples: lessons from the detection of salmonellae in pigs. J Clin Microbiol. 2005;43:3033–3037.
38. Schuurman T, de Boer RF, van Zanten E, van Slochteren KR, Scheper HR, Dijk-Alberts BG, Möller AVM, Kooistra-Smid AMD. Feasibility of a molecular screening method for detection of Salmonella enterica and Campylobacter jejuni in a routine community-based clinical microbiology laboratory. J Clin Microbiol. 2007;45:3692–3700.
39. Singer RS, Cooke CL, Maddox CW, Isaacson RE, Wallace RL. Use of pooled samples for the detection of Salmonella in feces by polymerase chain reaction. J Vet Diagn Invest. 2006;18:319–325.
40. Stone GG, Oberst, RD, Hays MP, McVey S, Chengappa MM. Detection of Salmonella serovars from clinical samples by enrichment broth cultivation-PCR procedure. J Clin Microbiol. 1994;32:1742–1749.
41. Bohaychuk VM, Gensler GE, McFall ME, King RK, Renter DG. A real-time PCR assay for the detection of Salmonella in a wide variety of food and food-animal matrices. J Food Protect. 2007;70:1080–1087.
42. Eriksson E, Aspan A. Comparison of culture, ELISA and PCR techniques for salmonella detection in faecal samples for cattle, pig and poultry. BMC Veterinary Research 2007;3:21. Available at: http://www.biomedcentral.com/1746-6148/3/21. Accessed 19 December 2008.
* Non-referred reference.