Patterson AR, Karriker LA, Evans RB, et al. Evaluation of alternative antemortem diagnostic samples for porcine reproductive and respiratory syndrome virus. J Swine Health Prod. 2007;15(6):339–345
| Original research | Peer reviewed |
Cite as: Patterson AR, Karriker LA, Evans RB, et al. Evaluation of alternative antemortem diagnostic samples for porcine reproductive and respiratory syndrome virus. J Swine Health Prod. 2007;15(6):339–345.
Also available as a PDF.
SummaryObjective: To assess the diagnostic accuracy of two minimally invasive methods of blood collection and a reference method. Materials and methods: Blood samples were collected from 30 pigs at 7 and 8 weeks of age. Fifteen pigs were then inoculated with porcine reproductive and respiratory syndrome virus (PRRSV) VR-2332 and 15 remained uninoculated. Pigs were sampled weekly for 7 weeks post inoculation (PI) using a reference sample (jugular vein sample) and two index samples (whole blood from the auricular vein collected either with a sterile polyester swab or using a capillary tube system). All samples were tested by quantitative reverse transcriptase-polymerase chain reaction (qPCR) and enzyme-linked immunosorbent assay (ELISA) using established protocols. Continuous sample data for the three sampling methods were compared by analysis of the area under the receiver-operating characteristic curve. Results: Sensitivity and specificity of qPCR testing for all samples ranged from 93% to 100% for weeks 1 through 3 PI. Results of ELISA testing depended on cutoff selection. Optimized ELISA sample:positive (S:P) ratio cutoffs for swab-sample data were significantly lower (mean S:P ratio cutoff = 0.08, SD = 0.05) than the industry standard (0.4). When the industry standard cutoff of 0.4 was utilized, swab-sample sensitivity ranged from 20.0% to 55.6% over weeks 2 through 7 PI. Implications: Diagnosis of viremic animals using qPCR can be equivalently accomplished using any of the sampling methods. PRRS ELISA status can be determined using any of the sampling methods if an alternative S:P ratio cutoff is used. | ResumenObjetivo: Evaluar la exactitud del diagnóstico de dos métodos minimamente invasivos de recolección de sangre y un método de referencia. Materiales y métodos: Se recolectaron muestras de sangre de 30 cerdos a las 7 y 8 semanas de edad. Se inocularon quince cerdos con el virus del síndrome reproductivo y respiratorio porcino (PRRSV por sus siglas en inglés) cepa VR-2332 y 15 no se inocularon. Durante 7 semanas post inoculación (PI por sus siglas en inglés) se tomaron muestras semanales de los cerdos utilizando una muestra de referencia (muestra de la vena yugular) y dos muestras índice (sangre completa de la vena auricular recolectada con un hisopo de poliéster estéril o utilizando un sistema de tubo capilar). Todas las muestras fueron probadas con la reacción en cadena de la transcriptasa reversa cuantitativa (qPCR por sus siglas en inglés) y la prueba de inmunoabsorbencia ligada a la enzima (ELISA por sus siglas en inglés) utilizando protocolos establecidos. Se compararon los datos de muestras continuas para los tres métodos de muestreo mediante el análisis del área bajo la curva característica de receptor operativo. Resultados: La sensibilidad y la especificación de la prueba qPCR para todas las muestras varió de 93% a 100% para las semanas 1 a 3 PI. Los resultados de la prueba de ELISA dependieron en la selección del punto de corte. El punto de corte óptimo para la relación muestra:positivo (S:P por sus siglas en inglés) de ELISA para las muestras de hisopos fueron considerablemente menores (punto de corte medio S:P promedio = 0.08, SD = 0.05) que el estándar de la industria (0.4). Cuando se utilizó el punto de corte estándar de la industria de 0.4, la sensibilidad de la muestra de hisopo varió de 20.0% a 55.6% de la 2 semana hasta la 7 PI. Implicaciones: El diagnóstico de animales virémicos utilizando el qPCR puede lograrse de manera equivalente utilizando cualquiera de los métodos de muestreo. El status de PRRS ELISA puede determinarse utilizando cualquiera de los métodos de muestreo si se utiliza un punto de corte S:P alternativo. | ResuméObjectif: Évaluer la précision diagnostique de deux méthodes de prélèvement de sang peu invasives et une méthode de référence. Matériels et méthodes: Des échantillons sanguins ont été prélevés sur 30 porcs à l’âge de 7 et 8 semaines. Quinze porcs ont par la suite été inoculés avec le virus du syndrome reproducteur et respiratoire porcin (PRRSV) VR-2332 et 15 sont demeurés non-inoculés. Les porcs ont été échantillonnés hebdomadairement pendant 7 semaines post-inoculation (PI) en utilisant un échantillon référence (échantillon de la veine jugulaire) et deux échantillons index (sang entier provenant de la veine auriculaire prélevé soit avec un écouvillon stérile en polyester ou un système utilisant un tube capillaire). Tous les échantillons étaient éprouvés par réaction d’amplification en chaîne quantitative utilisant la transcriptase réverse (qPCR) et une épreuve immuno-enzymatique (ELISA) utilisant des protocoles établis. Les résultats continus pour les trois méthodes d’échantillonnage ont été comparés par analyse de la surface sous la courbe caractéristique de la performance d’un test. Résultats: La sensibilité et la spécificité de l’épreuve qPCR pour tous les échantillons variaient de 93% à 100% entre les semaines 1 et 3 PI. Les résultats de l’épreuve ELISA dépendaient du seuil limite choisi. Les seuils limites optimisés pour l’épreuve ELISA du ratio échantillon:positif (S:P) pour les échantillons avec écouvillon étaient significativement inférieurs (seuil limite moyen du ratio S:P = 0.08, SD = 0.05) au standard de l’industrie (0.4). Lorsque le seuil limite de 0.4 était utilisé, la sensibilité de l’échantillon utilisant un écouvillon variait entre 20.0% et 55.6% pour les semaines 2 à 7 PI. Implications: L’identification d’animaux virémiques à l’aide de qPCR peut être accomplie de manière équivalente en utilisant n’importe laquelle des méthodes de prélèvement. Le statut quant au PRRS à l’aide de l’ELISA peut être déterminé en utilisant n’importe laquelle des méthodes d’échantillonnage si un seuil limite alternatif du ratio S:P est utilisé. |
Keywords: swine, porcine
reproductive and respiratory syndrome virus, PRRSV, antemortem diagnosis
Search the AASV web site
for pages with similar keywords.
Received: January
13, 2007
Accepted: July
23, 2007
Although porcine reproductive and respiratory syndrome (PRRS) has been recognized since 1987,1 diagnosis by clinical signs alone remains challenging, as many other viral and bacterial diseases have a similar clinical presentation. Therefore, diagnosing PRRS virus (PRRSV) infections and monitoring herd status has historically relied on laboratory testing, including virus isolation, immunohistochemistry, reverse transcriptase-polymerase chain reaction- (RT-PCR-) based assays, and serological tests such as enzyme-linked immunosorbent assays (ELISAs), immunofluorescence assays (IFAs), and the immunoperoxidase monolayer assay.2 While these diagnostic tests have well-described advantages and disadvantages,2 none is perfectly sensitive and specific.
Multiple factors influence performance of diagnostic tests. One factor is the composition of the submitted sample. Serum samples are commonly used for antemortem diagnosis and monitoring of PRRSV infection. Various sites can be used to collect blood,3 including ear veins, tail vessels, other peripheral vessels, the orbital sinus, and vessels in the thoracic inlet (jugular vein or anterior vena cava, herein referred to as the jugular sampling method). While the jugular sampling method remains commonplace, potential for injury to the thyroid gland, the phrenic nerve, and the thoracic duct has been noted.4-8 The jugular sampling technique also normally requires restraint of the animal with a wire snare. This manner of restraint has been used as a method of stress induction in various endocrine studies,9-11 indicating that it is stressful to the animal. Additionally, it is potentially hazardous to the human handler.
As diagnostic testing of blood samples for PRRS becomes an increasingly valuable marketing tool in boar studs, and remains a necessity for disease monitoring in herds, development and evaluation of an easier, less invasive, and safer method of blood collection is needed. One such method, pricking the ear with a needle and collecting blood with a sterile polyester swab, has recently gained popularity in boar studs.12-14 The whole blood collected is diluted in sterile saline or phosphate buffered saline. A recent study advocated the use of this method in PRRS-negative boar studs as a more appropriate PRRS-monitoring method than jugular sampling.14 While that study found swab samples to be a reliable alternative to the jugular sample, the trial monitored the use of swabs with quantitative PCR (qPCR) for only the first 6 days post PRRSV infection.14 This sampling method is prone to variation both in the amount of sample collected (Patterson AR, Karriker LA, Yoon KJ, unpublished data, 2007) and in subsequent dilution factors across samples. Significant variation in diagnostic accuracy of PRRSV qPCR testing is likely.15 Consequently, the diagnostic accuracy of the swab sampling method throughout the infectious period, especially during later stages of infection, must be assessed before this technique is applied to wider testing protocols. Additionally, diagnostic accuracy of swabs for ELISA testing has not been reported. The emergence of PRRSV strains that escape detection by some PCR methods (Dr Jim Collins, University of Minnesota, written communication, 2006) illustrates the urgent need for additional diagnostic approaches. Using the ELISA assay in preference to PCR for PRRS monitoring may provide more complete information as strain divergence occurs.
An alternative method of collecting blood from the ear uses a device in which a plastic capillary tube is coupled with a microcentrifuge tube containing serum separator gel or anticoagulant.15 This provides a serum sample, unlike the swab sample which provides diluted whole blood. The additional expense and labor involved with the microcentrifuge-tube method must be weighed against its potential advantages.
The goal of this study was to analyze the diagnostic accuracies of two standard diagnostic tests for PRRS when blood samples are collected using the polyester-tipped swab method, the capillary-tube method, and the jugular sampling method. Diagnostic tests included qPCR, used to identify viremic animals, and ELISA, used to identify antibody as an indication of exposure to PRRSV. As the sensitivity and specificity of the ELISA test depend on the sample:positive (S:P) ratio cutoff chosen to discriminate between seropositive and seronegative pigs, analysis was conducted to assess the optimal ELISA cutoff values for the study population.
Materials and methods
Pigs
Thirty 7-week-old, mixed-gender, crossbred, PRRSV-negative pigs (21.4 ± 1.4 kg; mean ± SE) were procured from a commercial herd which was considered PRRSV-negative on the basis of 2 years of regular monitoring by both ELISA and qPCR. The pigs were transported from the commercial operation to an Iowa State University research facility, individually tagged, and randomly assigned to two groups of 15 pigs upon arrival. Groups were housed separately in identical facilities, fed a commercial diet ad libitum, and cared for according to established criteria.16 The protocol for this trial was approved by Iowa State’s Institutional Animal Care and Use Committee.
Study design
Blood samples were collected from all pigs on arrival (Day 0) and approximately 1 week later (Day 6) to confirm PRRSV-negative status via IFA, qPCR, and ELISA (HerdChek PRRS 2XR ELISA, Idexx Laboratories, Westbrook, Maine). Upon confirmation of negative status, 15 pigs served as an uninfected control group. On Day 7, the remaining 15 pigs were inoculated intramuscularly with 2 mL of an inoculum containing 103 median tissue culture infectious doses of PRRSV (VR-2332).17,18
Blood samples were collected from all pigs once a week for 7 weeks post inoculation (PI). At each sampling time, blood was collected from each pig using three methods. One sample was collected from the jugular vein using a 9-mL serum separator tube (Vacutainer SST; BD, Franklin Lakes, New Jersey) and an 18-gauge, 1” needle. For the second method, an auricular vein was lanced with a 20-gauge, 0.5” needle. Blood was collected by saturation of a sterile polyester-tipped applicator (Product number 14-959-90; Fisher Scientific International, Waltham, Maine). The swab was placed in a 5-mL sterile polystyrene culture tube (Product number 352058, BD) with 1.0 mL sterile physiological saline (0.9% Sodium Chloride; Hospira, Lake Forest, Illinois). For the third method, an auricular vein was lanced with a 20-gauge, 0.5” needle, and a commercial capillary blood-collection system (SAFE-T-FILL Serum Gel Capillary Collection; RAM Scientific, Yonkers, New York) was used to collect approximately 200 μL of blood.
The order of sampling was randomized by arbitrarily selecting a sampling order for each pig from a box. This designated sampling order was then used for the remainder of the trial. Jugular and capillary-tube samples were centrifuged per manufacturers’ recommendations to separate the serum and blood cells. Blood swabs were processed according to a protocol reported by Chung et al 2005.19 Specifically, samples were vortexed for 15 seconds, then centrifuged at 4°C at 1738g for 10 minutes. Approximately 0.5 mL of supernatant was submitted for testing. All samples were refrigerated overnight and submitted the following morning to Iowa State University Veterinary Diagnostic Laboratory for qPCR and ELISA testing using established protocols.20,21 Technicians performing the qPCR and ELISA tests were blinded to the infection status of the animals and to trial objectives. Sample type was not blinded due to obvious visual differences between sample colors and containers. For PCR testing, a set of standards, each of which contained a known virus titer (fluorescent focus forming unit [FFU] per mL), was included in each qPCR run to determine the validity and reproducibility of the assay. The amount of PRRSV in each sample was estimated by converting the value for the threshold cycle (Ct; the cycle in which the PCR amplicon of the target genetic material is first detected19) to a virus titer (FFU per mL) using a standard curve.
Statistical analysis
Receiver operating characteristic (ROC) curve analysis was used to assess the diagnostic accuracy of the three sampling methods at each sampling point. This method utilizes the entire range of test data, providing a comprehensive overview of sample diagnostic accuracy independent of prevalence.22 Specifically, ROC curve analysis was utilized to select the cutoff values that optimized sample sensitivity and specificity at each sampling point, to use optimal cutoffs to analyze the diagnostic sensitivity and specificity of all samples at each sampling point, and to compare areas under the ROC curve (AUC) among samples at each sampling point. An AUC value is interpreted as the “probability that a randomly drawn individual from the positive reference sample has a greater test value than a randomly drawn individual from the negative reference sample.”23 An AUC of 0.80, for example, would indicate that 80% of the time, a randomly selected known PRRS-ELISA-positive pig will have an ELISA S:P ratio higher than that of a randomly selected known PRRS-ELISA-negative pig.22 Areas under the ROC curve range from 0.5 to 1, with 1 indicating that using a given sample, the diagnostic test would perfectly discriminate between PRRS-positive and PRRS-negative pigs, and 0.5 indicating that there is no discrimination between groups.22 Confidence intervals (95%) are given for AUC values. When the confidence interval includes 0.5, there is evidence that the test is not discriminating between groups.22
Pairwise comparisons of AUC values were analyzed for weekly samples to determine whether there were significant differences among sampling methods. Specifically, at each sampling point, the AUC value for the jugular sampling method was statistically compared24 to the AUC value for the swab method and a P value was generated; P values < .05 were considered significant. Similarly, a pairwise comparison of the jugular AUC value and the capillary sample AUC value was performed. ROC analysis was performed using standard statistical software (MedCalc Version 9.1.0.1; Mariakerke, Belgium).
Results
All 30 pigs remained enrolled for the duration of the trial and were monitored daily for lameness and clinical signs of respiratory disease. During the 9-week trial, a total of four pigs were treated either for respiratory conditions or lameness or both (two control and two infected animals), one pig for rectal prolapse (control animal), and one pig for a tail bite (control animal), according to recommendations by the university veterinarian. No adverse reactions, including ear hematomas persisting longer than 2 days post collection, were noted at the site of blood collection.
Results of qPCR testing (detection of viremia)
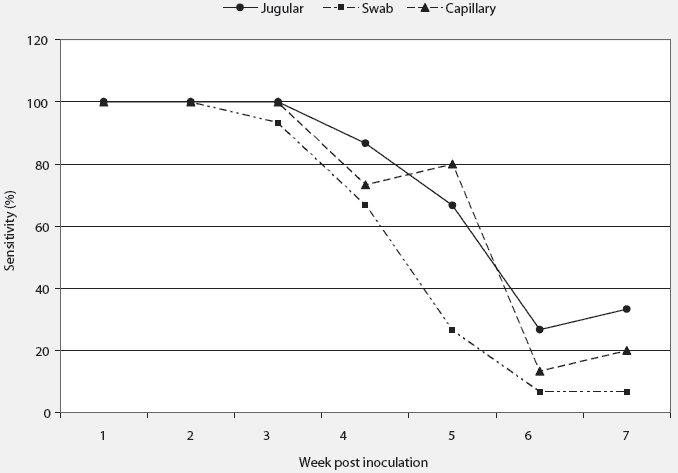
ROC curve analysis illustrated very high AUC values (very good discrimination) for all samples for the first 4 weeks PI (Table 1). After this time, test discriminating decreased for all sampling methods (Table 1). Pairwise comparison of AUC values showed significant differences only when capillary-tube and swab samples were compared on week 5 PI (P = .02). Optimization of Ct cutoff points for continuous qPCR data are presented in Table 2. The sensitivity of all sampling methods, calculated using optimized cutoff values, was high for weeks 1 to 3 PI, but decreased thereafter (Figure 1). Specificity of the sampling methods remained high throughout the testing period. The jugular sample specificity ranged from 93% to 100%, while specificity of the swab and capillary samples remained at 100% throughout the testing period.
Table 1: Summary of receiver operating characteristic (ROC) analysis of continuous data from samples collected by jugular, swab, and capillary-tube sampling methods and tested by real-time quantitative reverse-transcriptase polymerase chain reaction for PRRSV*
* Groups of pigs housed separately were either inoculated with porcine reproductive and respiratory syndrome virus (PRRSV) at 8 weeks of age (n = 15) or served as uninoculated controls (n = 15). All pigs were sampled weekly for 7 weeks post inoculation (PI) by three sampling methods. Blood samples were collected at weekly intervals PI by jugular venipuncture collected using a 9-mL serum separator tube (Vacutainer SST; BD, Franklin Lakes, New Jersey) or by lancing the auricular vein and collecting blood either by sterile polyester swab or by a capillary-tube system (SAFE-T-FILL Serum Gel Capillary Collection, RAM Scientific, Yonkers, New York). † ROC curves were significantly different (P = .02, pairwise ROC curve comparison24). |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 2: Optimized cutoff values* for qPCR for PRRSV and ELISA for antibodies to PRRS virus at each sampling point for the jugular, swab, and capillary blood-sampling methods†
* Receiver operating characteristic (ROC) curve analysis was used to determine the highest cutoff that provided the maximum sensitivity and specificity at each sampling point for qPCR and ELISA assays. † Pigs and sampling methods described in Table 1. ‡ Capillary samples were accidentally not submitted for ELISA testing on week 1 PI. PRRSV = porcine reproductive and respiratory syndrome (PRRS) virus; qPCR = quantitative reverse-transcriptase polymerase chain reaction; ELISA = enzyme-linked immunosorbent assay; PI = post inoculation; S:P ratio = sample-to-positive ratio; Ct = threshold cycle, ie, cycle in which the PCR amplicon of the target genetic material is first detected;19 ND = not done. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Figure 1: Diagnostic sensitivity of three sampling
methods for diagnosis of porcine reproductive and respiratory syndrome
virus (PRRSV) using real-time quantitative reverse transcriptase-polymerase
chain reaction (qPCR). Sensitivity was determined using receiver operating
characteristic (ROC) statistical analysis in which sensitivity data is
generated on the basis of optimized cutoff values for data dichotomization.23 Crossbred
barrows and gilts with an initial mean weight (± SE) of 21.4 ± 1.4
kg were first sampled at 7 weeks of age. Fifteen animals were inoculated
intramuscularly with PRRSV and 15 animals were uninoculated. All pigs were
sampled weekly for 7 weeks post inoculation using swab, capillary, and
jugular methods of blood collection. The overall shape of the graph is
consistent with the dynamic nature of sensitivity over the course of an
infection. Note the similarity among sampling methods at each time point.
|
Results of ELISA testing (detection of seropositive animals)
Sensitivity and specificity of swab samples were highly dependent on cutoff selection. When the industry standard cutoff S:P ratio (ie, 0.4) was utilized, sensitivity ranged from 20.0% to 55.6% over weeks 2 through 7 PI (Table 3). In contrast, jugular-sample sensitivity was 100% over weeks 2 through 7 PI, and capillary-sample sensitivity ranged from 93.3% to 100% over weeks 2 through 7 PI, when the industry standard cutoff S:P ratio was used.
Table 3: Diagnostic sensitivity and specificity of PRRS ELISA on samples collected using the swab sampling method* when a cutoff S:P ratio of 0.4 or an optimized cutoff S:P ratio was applied to dichotomize continuous data†
* Pigs and study design are described in Table 1. Samples were collected by lancing the auricular vein and collecting blood on a sterile polyester swab (product number 352058, BD) which was then placed in 1.0 mL of sterile physiological saline. The sample was vortexed for 15 seconds then centrifuged at 4°C at 1738g for 10 minutes. Approximately 0.5 mL of supernatant was submitted for ELISA testing. † Two cutoff S:P ratios were used to dichotomize the continuous ELISA data for each week PI: the current industry standard cutoff (0.4) and an S:P ratio generated by receiver operating characteristic (ROC) analysis. The cutoff generated by ROC analysis was optimized both for diagnostic sensitivity and specificity and varied by week PI. PRRS ELISA: enzyme-linked immunosorbent assay for antibodies to porcine reproductive and respiratory syndrome virus; S:P ratio = sample-to-positive ratio; PI = post inoculation. |
The above sensitivity and specificity results were significantly different when an optimized S:P ratio cutoff was used. Sensitivity for all sampling methods, generated using cutoffs optimized through AUC analysis, was 100% for weeks 2 through 7 PI. Specificity, calculated by AUC analysis, was 100% for weeks 3 through 7 PI. Specificity on week 2 PI was 100% and 86% for jugular and swab samples, respectively. Optimized S:P ratio cutoffs for continuous ELISA data were substantially lower (average cutoff = 0.07) for the swab sampling method than for either the jugular or capillary-tube sampling method (Table 2).
ROC curve analysis revealed that all sampling methods were able to perfectly distinguish (ie, AUC = 1) between known positive and negative animals for weeks 3 to 7 PI. Week 1 PI AUC values were 0.69 and 0.57 for the jugular and swab samples, respectively. Week 2 PI AUC values were 1 for jugular samples, 0.94 for swab samples, and 1 for capillary samples. Pairwise comparison revealed no significant differences among sampling methods at any time point.
Discussion
The problem of serum dilution (10-fold to 20-fold) when swab samples are submitted for PRRSV testing has been addressed in previous publications.13,14 This dilution likely results both from the inability to recover the entire sample from the swab and from placing the swab into sterile saline for transport. These and other unidentified factors result in smaller volumes of blood being submitted in swab samples. Additionally, when a swab sample is vortexed and centrifuged, the sample drawn off the top is not serum, as can be collected from the jugular sampling method, but contains remnants of red blood cells that may interfere with the ELISA test (Dr John Johnson, Iowa State University, oral communication, 2006). Therefore, when the swab is submitted to the laboratory and further diluted for ELISA testing (40:1 dilution is standard laboratory protocol), it is expected that a significantly smaller amount of antibody will be identified compared to that in a jugular serum sample. Further dilution also occurs when samples are pooled to decrease the cost of testing (eg, for PCR testing).
Because there is a lower antibody concentration in the swab sample, a lower cutoff point is necessary to adequately discriminate between positive and negative animals. The results of this study indicate that if a lower cutoff point is chosen (eg, S:P < 0.07 considered negative), swab samples will be diagnostically equivalent to jugular samples under conditions similar to those in this study for weeks 2 through 7 PI. Conversely, if the industry standard ELISA S:P cutoff of 0.4 is used to dichotomize data from the swab samples, there is a dramatic decrease in diagnostic accuracy, with the sensitivity of the test varying in this study from 20.0% to 55.6%, depending on time after inoculation. However, if a lower cutoff is selected when swab samples are submitted, the sensitivity of the test is 100% (under the conditions of this study) for all times when antibodies are expected to be present (weeks 2 through 7 PI). It is vital to understand that while this study provided evidence that the swab-sampling method can be used as an accurate test of previous exposure, it is accurate only when a lower ELISA S:P ratio cutoff is used to dichotomize results.
In contrast to the swab samples, capillary-tube samples suffer from neither inadequate volume, as they are not diluted in saline, nor differences in sample type, as serum can be collected. Therefore, as expected, this study indicated that the capillary sampling method can be used for ELISA testing with diagnostic accuracy equal to that for samples collected using the jugular sampling method (under trial conditions for weeks 2 through 7 PI) when the industry standard ELISA S:P ratio cutoff of 0.4 is used.
Comparison of real-time qPCR results indicated that testing samples collected by any of the sampling methods can provide accurate diagnosis in animals expected to be viremic (infected animals 1 to 3 weeks PI). This is in agreement with a previous report in which swab and serum samples (both tested by qPCR) were correlated for 6 days post infection.14 The data presented in this study indicate that late in the infection (weeks 5 through 7 PI), all three samples were relatively poor at detecting viremic animals. This can be explained by fully understanding the assumptions of the statistical model.23 For the purposes of AUC analysis, animals were considered positive or negative on the basis of initial inoculation status. As infection progresses, viremia is cleared and initial inoculation status no longer corresponds to infection status. Therefore, regardless of sample type, virus testing would likely be a less reliable indicator of infection than antibody testing as time from exposure elapses. After week 5 PI, regardless of sample type, qPCR was not predictive of infection in this study. Additionally, a significant difference (PÂ <Â .05 on a pairwise comparison)24 between the ROC curves on week 5 PI for swab and capillary tubes was noted. The difference of 20% in sensitivity between swab and jugular samples (when individual samples are tested at 5 weeks PI) should be carefully considered within the context of the testing situation, especially when the exact time of infection is unknown.
This study evaluated the diagnostic accuracy of three sampling methods for diagnosis of PRRSV by qPCR and ELISA testing, with diagnostic accuracy defined as “the ability to correctly classify subjects into clinically relevant subgroups.”22 Given this definition, the quality of information provided by the various samples, not their usefulness (or clinical practicality), was analyzed.22 In future studies, the usefulness of these sampling methods under field conditions should be assessed. Combining information from such a study with farm-specific economic and labor considerations will enable practitioners to develop effective PRRSV-testing protocols.
Implications
- Early diagnosis of PRRSV (1 to 3 weeks PI) using real-time qPCR can be equivalently accomplished using the blood-swab, jugular, or capillary methods of blood-sample collection.
- No change in cutoff values for qPCR data dichotomization is necessary for data obtained via any of the sampling methods.
- Under the conditions of this study, the diagnostic accuracy of the PRRSV ELISA is poor for blood samples collected using the swab method when the industry standard S:P ratio cutoff (0.4) is utilized.
- All sampling methods are capable of achieving very high diagnostic accuracy on PRRSV ELISA testing when optimal cutoffs (determined by AUC analysis) are employed, but these cutoffs may not be clinically useful.
References
1. Mengeling WL, Lager KM, Vorwald AC. Diagnosis of porcine reproductive and respiratory syndrome. J Vet Diagn Invest. 1995;7:3–16.
*2. Yoon K-J, Christopher-Hennings J, Nelson EA. Diagnosis. In: Zimmerman J, Yoon K-J, Neumann E, eds. 2003 PRRS Compendium. 2nd ed. Des Moines, Iowa: National Pork Board; 2003:59–68.
3. Muirhead M. Blood sampling in pigs. In Pract. 1981;3:16,18–20.
*4. Huhn RG, Osweiler GD, Switzer WP. Application of the orbital sinus bleeding technique to swine. Lab Anim Care. 1969;19:403–405.
5. Schwartz WL, Smallwood JE. Collection of blood from swine. NZ Vet J. 1977;25:237–238.
6. Lawhorn B. A new approach for obtaining blood samples from pigs. JAVMA. 1988;192:781–782.
7. Brown JR, Tyeryar EA. Femoral venipuncture for repeated blood sampling in miniature swine. Lab Anim Sci. 1978;28:339–342.
8. Bailie MB, Wixson SK, Landi MS. Vascular-access-port implantation for serial blood sampling in conscious swine. Lab Anim Sci. 1986;36:431–433.
9. Rushen J, Ladewig J. Stress-induced hypoalgesia and opioid inhibition of pigs’ responses to restraint. Physiol Behav. 1991;50:1093–1096.
10. Rushen J, Schwarze N, Ladewig J, Foxcroft G. Opioid modulation of the effects of repeated stress on ACTH, cortisol, prolactin, and growth hormone in pigs. Physiol Behav. 1993;53:923–928.
11. Farmer C, Dubreuil P, Couture Y, Brazeau P, Petitclerc D. Hormonal changes following an acute stress in control and somatostatin-immunized pigs. Domest Anim Endocrinol. 1991;8:527–538.
*12. Sanford SE. Breaking down the technical barriers to quick boar stud testing for PRRS. Better Pork. April 2005. Available at: http://www.betterfarming.com/bp/bp_2005/apr/apr05_stor3.htm#herd. Accessed 11 August 2007.
*13. Reicks DL. An overview of blood collection strategies for boar studs. Proc Allen D. Leman Swine Conf. St Paul, Minnesota. 2005;54–55.
14. Reicks DL, Munoz-Zanzi C, Rossow K. Sampling of adult boars during early infection with porcine reproductive and respiratory syndrome virus for testing by polymerase chain reaction using a new blood collection technique (blood-swab method). J Swine Health Prod. 2006;14:258–264.
*15. Yoon K-J, Patterson AR, Karriker LA. Alternatives to serum sampling for PRRS detection. Proc ISU Swine Dis Conf for Swine Pract. Ames, Iowa. 2005;139–140.
16. Federation of Animal Science Societies. Guide for the Care and Use of Agricultural Animals in Agricultural Research and Teaching. 1st ed. Savoy, Illinois: Federation of Animal Science Societies; 1999.
17. Yoon K-J, Zimmerman JJ, Chang C-C, Cancel-Tirado S, Harmon KM, McGinley MJ. Effect of challenge dose and route on porcine reproductive and respiratory syndrome virus (PRRSV) infection in young swine. Vet Res. 1999;30:629–638.
18. Rossow KD, Bautista EM, Goyal SM, Molitor TW, Murtaugh MP, Morrison RB, Benfield DA, Collins JE. Experimental porcine reproductive and respiratory syndrome virus infection in one-, four-, and 10-week-old pigs. J Vet Diagn Invest. 1994;6:3–12.
19. Chung W-B, Chan W-H, Chaung H-C, Lien Y, Wu C-C, Huang Y-L. Real-time PCR for quantitation of porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 in naturally-infected and challenged pigs. J Virol Methods. 2005;124:11–19.
20. Opriessnig T, McKeown NE, Harmon KL, Meng XJ, Halbur PG. Porcine circovirus type 2 infection decreases the efficacy of a modified live porcine reproductive and respiratory syndrome virus vaccine. Clin Vaccine Immunol. 2006;13:923–929.
*21. Idexx HerdChek PRRS 2XR [package insert]. Westbrook, Maine: Idexx Laboratories; 2006.
22. Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine [published erratum appears in Clin Chem 1993;39:1589]. Clin Chem. 1993;39:561–577.
23. Greiner M, Pfeiffer D, Smith RD. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev Vet Med. 2000;45:23–41.
24. Hanley JA, McNeil BJ. A method of comparing the areas under the receiver operating characteristic curves derived from the same cases. Radiology. 1983;148:839–843.
* Non-refereed references.