Original research |
Peer reviewed |
Prevalence and serovars of Salmonella enterica isolated from ileocolic lymph nodes of market pigs reared in selected Midwest US swine herds
Prevalencia y serovariedades de Salmonella entérica aislada de los nódulos linfáticos ileocólicos de cerdos de mercado criados en piaras seleccionadas del medio oeste de EU
Prévalence et sérovars de Salmonella enterica isolés des noeuds lymphatiques iléocaecaux de porcs élevés sur des fermes dans le midwest américain
Peter B. Bahnson, DVM, PhD; Daniel J. Damman, DVM; Richard E. Isaacson, PhD; Gay Y. Miller, DVM, PhD; Ronald M. Weigel, PhD; H. Fred Troutt, MS, VMD, PhD
PBB: Department of Medical Sciences, School of Veterinary Medicine, University of Wisconsin-Madison, Madison, Wisconsin. DJD, GYM, RMW: Department of Veterinary Pathobiology, College of Veterinary Medicine, University of Illinois at Urbana-Champaign, Urbana, Illinois. Dr Damman’s current affiliation is Indiana Animal Health, LLC, Wolcott, Indiana. REI: Department of Veterinary Pathobiology, College of Veterinary Medicine, University of Minnesota, St Paul, Minnesota. HFT: Department of Veterinary Clinical Medicine, College of Veterinary Medicine, University of Illinois at Urbana-Champaign, Urbana, Illinois. Corresponding author: Dr Peter Bahnson, Department of Medical Sciences, School of Veterinary Medicine, University of Wisconsin-Madison, 2015 Linden Drive, Madison, WI 53706; Tel: 608-265-1855; Fax: 608-265-8020; E-mail: pbbahnson@wisc.edu.
Cite as: Bahnson PB, Damman DJ, Isaacson RE, et al. Prevalence and serovars of Salmonella enterica isolated from ileocolic lymph nodes of market pigs reared in selected Midwest US swine herds. J Swine Health Prod. 2006;14(4):182-188.
Also available as a PDF.
SummaryObjectives: To describe the prevalence and serovars of Salmonella enterica in ileocolic lymph nodes of slaughtered swine in a sample of Midwest US herds and to assess as methods of study pooling and freezing of lymph-node samples prior to bacterial culture. Materials and methods: Ileocolic lymph nodes from 30 pigs from each of 146 herds were sampled at slaughter. Tissue from five pigs was pooled for one bacterial culture. Retained frozen tissues from the same pigs were cultured individually (n = 82 herds) from a subset of those with Salmonella-positive pools (n = 100 herds). A mathematical relationship was described to predict approximate individual prevalence based on number of positive pools. Isolates were serotyped. To test for effects of freezing on test sensitivity, lymph nodes from 100 pigs were cultured both fresh and after freezing. Results: Salmonellae were detected in 100 of 146 herds (68.5%). The mean number of positive pools per herd was 1.75, and the mean within-herd, individual-pig prevalence was 6.98% (95% CI, 4.88% – 9.07%). Freezing of samples did not result in decreased detected prevalence. Individual prevalence could be approximately predicted by pool results, although with low precision. Implications: Salmonellae were found in two-thirds of the herds studied. Culture of pooled samples with subsequent culture of retained frozen tissues from positive pools may be an effective way to test a larger number of herds on a given budget through laboratory-cost savings. However, pooling without culture of individuals from positive pools results in imprecise prevalence estimation. | ResumenObjetivos: Describir la prevalencia y serovariedades de Salmonella entérica en nódulos linfáticos ileocólicos de cerdos de mercado en una muestra de piaras del medio oeste de EU y valorar cólmo métodos de estudio el agrupamiento (pooling) y la congelacióln de muestras de nóldulos linfáticos antes del cultivo bacteriano. Materiales y métodos: Al sacrificio se tomaron muestras de los nóldulos linfáticos ileocóllicos de 30 cerdos de cada una de las 146 piaras. Se agrupól el tejido de cinco cerdos para un cultivo bacteriano. Los tejidos congelados de los mismos cerdos se cultivaron individualmente (n = 82 piaras) para un subgrupo de muestras agrupadas que resultaron positivas a Salmonella (n = 100 piaras). Se describiól una relacióln matemática para predecir la prevalencia individual aproximada basada en el número de agrupamientos positivos. Los aislamientos fueron serotipificados. Para probar el efecto de la congelacióln en la sensibilidad de la prueba, se cultivaron los nóldulos linfáticos de 100 cerdos, tanto frescos como después del congelamiento. Resultados: Se detectól la salmonellae en 100 de las 146 piaras (68.5%). El número promedio de agrupamientos positivos por piara fue de 1.75, y el promedio de la pre-valencia individual por cerdo dentro de la piara, fue de 6.98% (95% CI, 4.88% – 9.07%). El congelamiento de las muestras no disminuyól la prevalencia detectada. La prevalencia individual podría predecirse aproximadamente con los resultados del agrupamiento, aunque con poca precisióln. Implicaciones: Se encontról salmonellae en dos tercios de las piaras estudiadas. El cultivo de las muestras agrupadas con el cultivo subsecuente de los tejidos congelados de los agrupamientos positivos puede ser una manera efectiva de probar un número mayor de piaras con un presupuesto pre-establecido a través de un ahorro en los costos de laboratorio. Sin embargo, el agrupamiento sin el cultivo de muestras individuales de los agrupamientos positivos resulta en una estimacióln imprecisa de la prevalencia. | ResuméObjectifs: Déterminer la prévalence et les sérovars de Salmonella enterica isolés des noeuds lymphatiques iléocaecaux de porcs charcutiers abattus provenant d’un échantil-lonnage de fermes du midwest américain et évaluer comme méthode d’étude le regroupement et la congélation d’échantillons de nœuds lymphatiques avant de les soumettre à la culture bactérienne. Matériels et méthodes: Des nœuds lymphatiques iléocaecaux ont été prélevés à l’abattoir sur 30 porcs de chacune de 146 fermes. Les tissus provenant de cinq porcs ont été regroupés pour effectuer une seule culture bactérienne. Les tissus conservés congelés de ces mêmes animaux ont été cultivés individuellement (n = 82 élevages) pour obtenir un sous-groupe des fermes ayant des pools positifs pour la présence de Salmonella (n = 100 élevages). Une relation mathématique basée sur le nombre de pools positifs a été décrite pour prédire la prévalence individuelle approximative. Le sérotypage des isolats a été fait. Afin de vérifier les effets de la congélation sur la sensibilité du test, des nœuds lymphatiques provenant de 100 porcs ont été cultivés frais et après congélation. Résultats: La présence de salmonellae a été détectée dans 100 des 146 troupeaux (68.5%). Le nombre moyen de pools positifs par troupeau était de 1.75, et la moyenne intra-troupeau de la prévalence individuelle était de 6.98% (CI 95%, 4.88% – 9.07%). La congélation des échantillons ne causa pas de diminution de la prévalence détectée. Les prévalences individuelles pouvaient être approximativement prédites à partir des résultats des pools, quoique avec une faible précision. Implications: La présence de salmonellae été détectée dans le deux tiers des troupeaux étudiés. La culture des échantillons regroupés avec culture subséquente des tissus congelés provenant des pools positifs semble un moyen efficace pour tester un plus grand nombre de troupeaux pour un budget donné en réalisant des économies sur les analyses de laboratoire. Toutefois, le regroupement sans culture des échantillons individuels constituant les pools positifs donne une estimation imprécise de la prévalence. |
Keywords: swine, Salmonella prevalence,
microbiologic methods, epidemiology
Search the AASV web site
for pages with similar keywords.
Received: February
16, 2005
Accepted: July
11, 2005
Salmonella enterica is an important foodborne pathogen of pork. The US Centers for Disease Control and Prevention (CDC) estimated that nontyphoid Salmonella was associated with 16.1 cases per 100,000 population in 2002.1 The CDC also has estimated that there were more than 1.4 million human Salmonella cases per year in the United States, resulting in 15,000 hospitalizations and 400 deaths annually during 1996-1999.2 A survey of retail US pork products, both whole muscle and ground product, indicated that 9.6% of 384 tested products were positive for Salmonella serovars,3 while 1.6% of 613 pork chops collected at retail US markets in 2002 were Salmonella-positive.4
While the proportion of human cases directly or indirectly attributable to pork has been incompletely quantified in the United States, one report estimates that, on the basis of an assessment of outbreak data, 6% to 9% of foodborne Salmonella infections in the United States may be associated with pork and pork products.5 Although data is not directly transferable to the United States situation because of differences in consumption patterns and other factors among countries, reports from two other countries also suggest a role for pork in human salmonellosis. A study of Danish surveillance data estimated that 9% of human salmonellosis was attributable to pork, with 75% of all cases attributed to a source.6 Approximately 20% of human salmonellosis in Germany may be attributable to pork sources.7 Salmonella causes more severe illness than other common foodborne pathogens in pork, with relatively high hospitalization and death rankings for nontyphi Salmonella.2 Human infections by nontyphi Salmonella has a substantial economic impact in the United States,8 estimated at $US 3B.
Foodborne outbreaks of all types attributable to pork have declined in the United States during the period 1973 to 1992.9 Since 1992, US slaughter plants have instituted process control systems based on Hazard Analysis Critical Control Point (HACCP) principles to improve pork food safety. Salmonella serovars are being monitored by the United States Department of Agriculture (USDA) as an indicator of the success of these plans, and a performance standard specifying a maximum of 8.7% Salmonella-positive carcasses has been enforced since 1998.10 While the USDA has documented progress in Salmonella control, 2.5% of carcasses tested Salmonella-positive in 2003.11 Consequently, Salmonella in pork continues to represent a threat to food safety.
We designed this study to quantify the prevalence and serovars of Salmonella among slaughtered pigs from US Midwest commercial swine herds. To better understand and interpret our results, we also assessed the effect of freezing ileocolic lymph-node samples prior to culture for Salmonella isolation, and described the relationship between pooled and individual-sample culture results.
Materials and methods
Study herds
Two major US Midwest slaughter plants agreed to participate in the study on condition of anonymity. The client lists of these plants were reviewed to identify herds routinely capable of delivering at least 30 pigs per lot marketed on a single day. In addition, a state pork-producer association, coordinated marketing groups, and swine-dedicated veterinary practices provided names of herds likely to market to these slaughter plants. Participation request forms were distributed to 333 herds, and 225 agreed to participate (67.6%). The selection criteria for participation in the study were that the herds sold market weight pigs to abattoirs participating in the study, were located in the same state as the slaughter plants, were able to market at least 30 animals as a single group, and agreed to complete a survey. Herds delivered pigs to the slaughter plant on the schedule of their choice. Pigs were sampled on the basis of availability of technical personnel at the time of delivery. Samples were collected between July 17, 1997 and October 9, 1998, with a target to sample up to 150 herds. Herds were selected without prior knowledge of Salmonella occurrence.
Collection of ileocolic lymph nodes
Pigs were transported to slaughter using the herd’s normal delivery methods, and were placed in lairage for variable lengths of time, as was the practice of the slaughter plants. Pigs were marked with a tattoo, segregated into a holding pen, and moved through the slaughter line as a group. For lots with more than 30 animals, only the first 30 animals on the slaughter line were collected. After evisceration, the intestinal tract was removed from the slaughter line, and the tracts were placed in collection bins. Care was taken not to place tracts from more than one herd in any bin. These were moved to a separate area for removal of the ileocolic lymph nodes (lymph nodes draining the cecum, ileum, and colon, also commonly referred to as ileocecal lymph nodes). The overlying mesentery was wiped dry with sterile gauze, then carefully reflected using sanitized instruments to prevent contamination of lymph nodes from the surface of the mesentery. The nodes were grasped using sterile gauze held by a clean gloved hand, collecting a sample expected to weigh > 4 g per individual and placing each in a separate sterile plastic sample bag. Samples were transported to the laboratory on ice.
Identification of Salmonella serovars in fresh samples
Salmonellae were identified using a modification of a published method.12 All inoculated media were incubated at 37°C. Within 24 hours of collection, lymph-node tissues from each individual were split approximately in half using a sterile scalpel or scissors. One half was placed in a sterile plastic bag and frozen at -70°C for up to 14 months. Two-gram samples from the second (unfrozen) halves from five pigs were combined, broken down with a mallet and a paddle blender (Stomacher 80; Seward Ltd, Worthington, UK), blended with 90 mL of tetrathionate broth (Tetrathionate Broth Base; Remel Inc, Lenexa, Kansas), and incubated for 42 to 48 hours. After incubation, 1 mL was added to a tube of 9 mL of R10 broth (Rappaport-Vassiliadis R10 Broth; Remel Inc) and incubated for 24 hours. Broths were streaked for isolation on XLT4 agar (XLT4 Agar Base; Remel Inc). If suspect colonies were found after 24 hours of growth, one colony was transferred to brilliant green agar. Suspect colonies were tested for agglutination with anti-Salmonella antibodies (Polyvalent O groups A through G Salmonella Somatic Agglutinating Serum, Rabbit; Remel Inc). Isolates failing agglutination were further tested using a battery of biochemical tests in a commercially available test kit (API 20E; bioMerieux, Hazelwood, Missouri). Isolates positive either by agglutination or the test kit were serotyped either at the US Department of Agriculture National Veterinary Services Laboratory (Ames, Iowa) or the Wisconsin State Veterinary Diagnostic Laboratory (Madison, Wisconsin).13
Identification of Salmonella serovars in frozen samples
Frozen samples were stored for testing at a convenient time, with the number of samples stored limited by available freezer space. Frozen lymph-node tissue from individuals in Salmonella-positive pools was thawed overnight at 2°C and cultured using the laboratory process described for fresh samples, except that 2 g of lymph node from each individual was blended with 20 mL tetrathionate broth and cultured separately.
Participating herd survey
A survey was mailed to herd managers the day after collection of samples including questions on facilities, husbandry, management, and slaughter-transport practices. Farm demographics are reported here; analysis to identify risk factors for prevalence of antimicrobial-resistant Salmonella are reported elsewhere.14 Nonrespondents were initially sent a reminder card and subsequently a second copy of the survey if they did not reply to the reminder. Finally, nonrespondents were contacted by telephone.
Statistical analysis
Pooled-sample prevalence. Potential differences in the distributions of pooled-sample results between the two slaughter plants was assessed using the Wilcoxon-Mann-Whitney test, which treats the number of positive pools as ordered categorical variables (StatXact, 4.0.1; Cytel Software Corporation, Cambridge, Massachusetts).
Individual-pig prevalence. Pig-level prevalence was calculated as the number of (frozen) individual-pig culture-positive samples divided by the total number of samples. Individuals with negative pool results were assumed to be Salmonella-negative, although these samples were not cultured individually. To estimate individual prevalence in herds with only pooled-sample results, a regression of individual pig prevalence as a function of the number of positive pools per herd was used to derive a statistical model (Statistica version 6.1; Statsoft Inc, Tulsa, Oklahoma). The square-root transformation of prevalence was used to meet the statistical model assumptions required. Overall prevalence was estimated by averaging across herds from individual cultures, where available, or from the predictive equation where samples were not individually cultured.
Preliminary testing for detection concordance. To assess the potential for freezing to bias culture results, we collected > 4 g of ileocolic lymph-node tissue from each of 100 carcasses at one slaughter plant. The samples were collected in four collections spread over an 8-hour work shift on a single day, sampling every fifth carcass on the slaughter line during each collection. Upon arrival at the laboratory, 4 g of tissue was weighed, and each node was cut in half. Half of each node was immediately processed. The remainder was placed in a sterile plastic bag, frozen at -70°C for 2 weeks, and then thawed overnight at 2°C. Culture methods for both fresh and frozen tissue were as described for individual samples, except that isolates were not serotyped. Detection concordance was compared by the kappa statistic using exact methods (StatXact).
Results
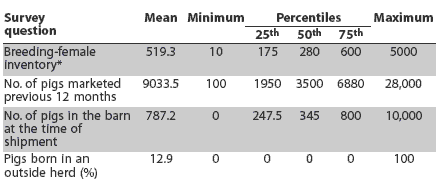
Of the 225 herds that agreed to participate, 146 herds were sampled and surveys were sent to them. Valid surveys were returned by 113 herds. Of these, 20 (17.7%) obtained some or all growing pigs from at least one outside herd, with the remaining herds rearing all pigs from birth to slaughter. The breeding-female inventory, number of pigs marketed during the previous 12 months, growing-pig inventory in the barn at the time of shipment, and proportion of piglets born in an outside herd were described (Table 1). Batch or all in-all out pig flow was practiced in the finisher barn by 39.4% of herds. Pigs were shipped directly from the farm to the slaughter plant by 104 herds (92.0%); four (3.5%) delivered to a facility where pigs from multiple herds were grouped for shipment to slaughter; and five (4.4%) reported other delivery methods.
Table 1: Summary of responses to herd survey questions among 113 Midwest swine herds
* For the 94 herds with breeding stock. |
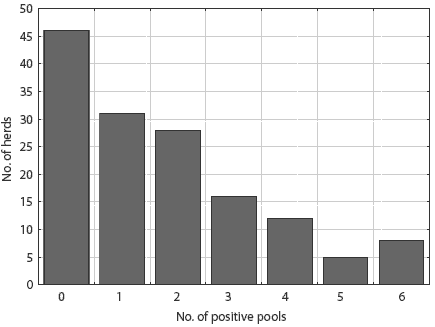
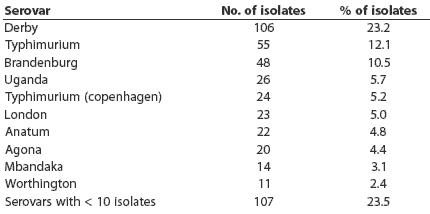
Salmonellae were detected in lymph nodes from 100 of 146 herds (68.5%). The distribution of pooled-sample results did not differ between slaughter plants (P = .71). The number of pooled positive samples varied from zero to six of six pools tested per herd, with a mean of 1.75 positive (Figure 1). Thirty-three serovars were detected. Ten serovars were represented by more than ten isolates (Table 2): these serovars accounted for 76.5% of all isolates. The other serovars detected were 4,5,12:i-mono-phasic, Bareilly, Bovis-morbificans, Bredeney, Chailey, Cholerasuis (Kunz), Cubana, Give, Hartford, Heidelberg, Infantis, Johannesburg, Litchfield, Livingstone, Montevideo, Muenchen, Newport, Ohio, Pakistan, Saint-Paul, Schwarzengrund, Tennessee, and Thompson.
Figure 1: Frequency distribution of 146 swine herds by the number of Salmonella culture-positive pools of ileocolic lymph nodes among six pools collected (five pigs sampled per pool).
|
Table 2: Salmonella enterica serovars detected among ileocolic lymph node samples from slaughtered pigs from 113 Midwest herds*
* Ileocolic lymph nodes were collected from 30 pigs per herd. Half of each individual pig sample was frozen. The other halves of individual fresh samples were pooled and cultured for Salmonella (five pigs per pool, six pools per herd). For Salmonella-positive pools, individual frozen samples were cultured and Salmonella isolates serotyped. |
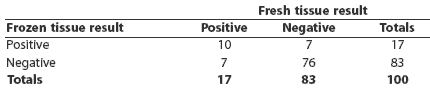
Of the 100 samples cultured both as fresh and frozen tissues, a total of 24 were culture-positive by at least one method, and 10 were positive by both methods (Table 3). Prevalence estimated by both methods was the same, although seven individuals that were positive by each method were negative by the other. The kappa statistic was 0.50 (95% CI, 0.28 – 0.73).
Table 3: Salmonella culture results from 100 paired ileocolic lymph-node tissues collected at slaughter and cultured fresh (not frozen) and after storing at -70 degrees C for 14 days (frozen)
|
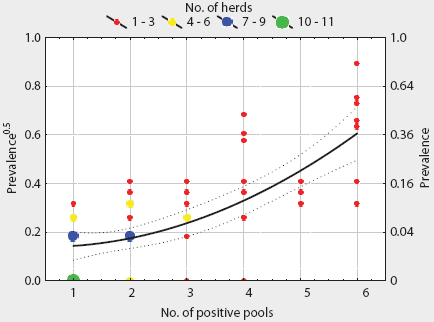
A total of 1005 frozen individual lymph-node samples were cultured from 201 positive pools distributed among 82 herds. These samples were frozen for 2 to 14 months, depending on availability of lab resources. Among these herds, the median within-herd prevalence of positive individuals was 6.67% (95% CI, 6.73% – 13.20%) and the mean was 9.96% (95% CI, 0% – 80.0%). No salmonellae were detected among individual cultures from 22 of these herds, although salmonellae were isolated from pooled samples. At the herd level, the number of positive pools was positively correlated with individual prevalence (r = 0.65; P < .01). Transformed individual prevalence was described by a quadratic equation (Table 4) derived by regression analysis. The fitted within-herd prevalence varied from 2.1% (95% CI, 0.7% – 4.0%) for herds with one of six positive pools to 36.8% (95% CI, 24.7% – 51.1%) for herds with six of six pools positive (Figure 2). The distribution of number of Salmonella-positive pools among pool-positive herds was not significantly different in herds with or without individual culture results (P = .78). The mean prevalence for all herds was estimated using frozen tissue for 82 herds and the predictive equation for the 22 culture-positive herds without individual results, and was set at zero for herds with no positive pools. The estimated mean prevalence was 6.98% (95% CI, 4.88% – 9.07%) and the 10th, 25th, 50th, 75th, and 90th percentiles were 0%, 0%, 2.5%, 7.0%, and 20%, respectively, with a range of 0% to 80%.
Table 4: A regression model of Salmonella prevalence0.5 in ileocolic lymph nodes of slaughtered pigs as a function of the number of culture-positive pooled samples*
* Samples from five individuals were combined to form a single pool for bacterial culture (six pools per herd) as described in Table 2 |
Figure 2: Salmonella prevalence in ileocolic lymph nodes of swine, modeled as a function of the number of culture-positive pooled samples per herd (six pooled samples per herd, 82 herds). Approximate 95% CIs for the fitted model are shown. Lymph nodes were collected at slaughter and stored at -70 degrees C for 2 to 14 months before culturing.
|
Discussion
Approximately two-thirds of the herds investigated in this study had at least one Salmonella culture-positive result, suggesting the importance of procedures to minimize pork contamination throughout the pork chain. These findings are similar to results of a study of Minnesota herds,15 where Salmonella were detected in 16 of 25 farms and in 3.6% of 3442 pigs. The Minnesota study differed from this study in culture and collection methods, and in addition, a variable number of pigs per herd were sampled in the Minnesota study (n = 14 to 1172). A survey of 317 pigs in Canadian slaughter facilities during 1985-1986 reported 14.2% positive (mesenteric lymph- node samples).16 Studies of five North Carolina farms17 and six Iowa farms18 reported 21% and 9.15% culture-positive pigs, respectively (ileocolic lymph-node samples). In Europe, findings of large-scale, population-based slaughter studies include 3.3% positive (n = 11,942) in Germany19 and 15.2% positive (n = 7756) in the Netherlands20 (ileocolic lymph node samples). In Denmark, 6.2% of cecal content samples were culture-positive (n = 13,468).21
The herds in this study were not selected in a manner that would ensure that they are representative of midwestern US herds. However, participants were chosen without knowledge of prior or current Salmonella status, farm management, or other herd characteristics other than size. Neither of the participating slaughter processors had specific programs linked to herd Salmonella history or current status of herds. Although the smallest herds were excluded from the study, since they were not able to deliver 30 pigs per shipment, most Midwest slaughter pigs are produced in herds at least the size of the study herds. Assuming weekly delivery, this minimum delivery-group size would correspond to a herd size of approximately 100 sows, assuming average production efficiency calculated from National Agricultural Statistical Service (NASS) figures.22 Midwest herds with current total inventory of 1000 pigs or larger, the NASS category closest to the inventory expected for 100 sows, accounted for 70.4% of all Midwest inventories in 1998.22 Only 10% of herds had ³ 20% prevalence, suggesting the possibility that on-farm interventions might be targeted to a subset of farms. However, the success of such a program would depend on development and implementation of tests that are both cost effective and able to accurately predict high risk of Salmonella shedding at slaughter. Further, the current study represents a one-time snapshot of prevalence. Since the prevalence of Salmonella may vary over time,23-25 identification of marketed groups with high prevalence may require ongoing sampling, which would be expensive and cumbersome using existing methods.
Regional differences have been reported for Salmonella prevalence based on fecal culture, with higher proportions of southeastern US herds positive compared with the herds in the Midwest and other areas.26 Consequently, it is likely that the results reported here are lower than would be found nationwide, assuming that fecal and ileocolic lymph-node detection rates are positively associated.27
The serovar distribution in swine in this study differed from that of isolates reported for human salmonellosis by the CDC’s FoodNet surveillance system.28 Most notably, the proportion of serovar Derby isolates was much higher in study samples compared with human-origin isolates. This discrepancy, among others, has been the basis for speculation that either pork may be a minor source of human salmonellosis in the United States, or that certain serovars, especially serovar Derby, have relatively low infectivity for humans.29 Serovar Derby was the most commonly detected serovar both in this study and in carcass swabs collected nationwide.11 On the other hand, there are commonalities among serovars isolated from pigs and human. First, eight of the 10 most common human serovars reported by the FoodNet system in 199828 were detected in swine lymph nodes in this study. Second, more than half of isolates from both swine and human FoodNet isolates were common serotypes. Whereas these commonalities do not prove that pigs are a source for human infection, the findings are consistent with such linkage and indicate the need for further study.
Invasive Salmonella infections result in more serious health consequences. Four of the eight most common invasive serovars reported in human disease30 were detected in swine samples in this study, although three of these, Heidelberg, Schwarzengrund, and Choleraesuis, were not among the 10 most common swine serovars detected in this study.
Samples collected at slaughter may reflect bacteria derived from the farm of origin, or those acquired during transportation and lairage. Although farms are an important source of strains isolated at slaughter,27 rapid Salmonella enterica infection of intestinal lymph nodes can occur,31,32 and there is evidence for uptake of new strains during transport and lairage.18 Consequently, isolates collected at slaughter should be regarded as a composite of all three sources.
These findings probably underestimate the true prevalence of Salmonella for several reasons, including the effect of pooling samples, freezing samples, and limited sensitivity of the culture methods used. Pooling followed by culturing of individual samples in positive pools was an effective way to reduce laboratory resources required. In this study, the total number of bacterial cultures was reduced by approximately two-thirds, compared with culturing all 30 individuals from every herd. Because the cost of culture media needed for Salmonella isolation is substantial for large-scale studies, this efficiency and cost savings can dramatically increase the number of herds and pigs studied. For epidemiologic studies, inclusion of a large number of herds provides for a more robust description of a population. However, pooled samples were not a precise indicator of individual-pig prevalence, suggesting that a two-stage protocol would help define prevalence more accurately. Pooling has the potential to introduce systematic bias if the culture methods for pools are less sensitive than culture methods for individual samples. An assessment of prevalence among samples paired with culture-negative pools would be useful to help interpret results. However, we did not evaluate this potential effect, since the objective of pooling in this study was to make more efficient use of available laboratory and financial resources, and culture of negative pools would have required resources that were instead used for study of additional pigs and herds. However, if this methodology resulted in lower sensitivity at the pig level, as would be expected, a bias toward underestimation of prevalence is the likely result. A further bias was noted in failure to detect Salmonella in individual samples paired to 22 of the positive pools. If it is assumed that none of the pooled-sample results were false-positives, then it follows that using individual results where they were available, as was done in this study, would also tend to underestimate prevalence. Possible reasons for these incongruities include potentially heterogeneous distribution of salmonellae in lymph-node tissues, relatively insensitive culture methods, death of bacteria during storage, and potential cross-contamination of pooled tissues, among others. These limitations should be kept in mind when pooled samples are used.
Freezing samples may result in diminished viability of salmonellae, with a consequent bias to underestimate prevalence in frozen samples, as has been reported in food samples,33 swine feces,34and poultry-house environmental samples.35 The finding of identical prevalence estimates in fresh and frozen samples, however, suggests that this effect, if present, appears to be fairly small for lymph-node cultures using the methods reported here. Salmonellae are believed to survive intracellularly in lymph-node tissue,36,37 and it is possible that enclosure of the organisms in host cells protected them from some of the damaging effects of freezing. The high number of discordant samples and moderate kappa statistic38 may suggest moderate to low sensitivity for the culture methods used, inconsistent distribution of salmonellae within lymph-node tissue, or both. Although the sensitivities of the specific methods used in this study have not been documented, other conventional methods applied to fecal samples have shown variable sensitivity, and in some cases, low sensitivity.39,40
While salmonellae were found in 68% of herds, the median prevalence of 6.7% suggests that in most herds, the proportion of culture-positive pigs was low. However, positive pigs pose a likely risk to pork food safety, since many of the serovars detected are also detected in human infection, and four were among the common list of invasive human serovars. These findings suggest the need for continued care in development of pork-chain Salmonella control programs, and the need for further research to identify cost-effective methods to reduce Salmonella shedding on farms.
Implications
- Under the conditions of this study, salmonellae were commonly isolated from ileocolic lymph nodes of slaughtered pigs, with positive results in approximately two-thirds of herds and an average of 7% of individual pigs.
- Among herds studied, a minority of market deliveries provided a disproportionately high prevalence of Salmonella-positive pigs.
- Freezing of lymph-node samples before bacterial culture appears an effective way to simplify logistics without compromising the ability to detect Salmonella.
- Pooling of samples for microbial culture then culturing individual retained (frozen) tissue from positive pools can be an effective way to reduce study costs, increase the number of herds examined, or both.
Acknowledgments
Financial support was provided by the Illinois Council on Food and Agricultural Research, the US Department of Agriculture, National Research Initiative, and the University of Wisconsin-Madison. The authors thank Xi-Li Li, Latifa Omran, and Lynn Volk for laboratory work.
References
1. Preliminary FoodNet data on the incidence of foodborne illnesses – selected sites, United States, 2002. Morbidity and Mortality Weekly Report. Weekly April 18, 2003. MMWR Morb Mortal Wkly Rep [serial online]. 2003;52(15). Available at: http://www.cdc.gov/mmwr/PDF/wk/mm5215.pdf. Accessed April 17, 2006.
2. Voetsch AC, Van Gilde TJ, Angulo FJ, Farley MM, Shallow S, Marcus R, Cieslak PR, Deneen VC, Tauxe RV. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis. 2004;38(Suppl 3): S127-134.
3. Duffy EA, Belk KE, Sofos JN, Bellinger GR, Pape A, Smith GC. Extent of microbial contamination in United States pork retail products. J Food Prot. 2001;64:172-178.
4. US Food and Drug Administration. Center for Veterinary Medicine. NARMS Retail Meat Annual Report, 2002. Table 6. Salmonella by serovtype and meat type, 2002. Available at: http://www.fda.gov/cvm/2002_RetailMeatRpt.htm. Accessed April 17, 2006.
*5. Frenzen PD, Buzby JC, Roberts T. An updated estimate of the economic costs of human illness due to foodborne Salmonella in the United States. Proc 3rd Int Symp Epidemiol Cont Salmonella Pork. 1999;215-219.
6. Hald T, Vose D, Wegener HC, Koupeev T. A Bayesian approach to quantify the contribution of animal-food sources to human salmonellosis. Risk Anal. 2004;24:255-269.
7. Steinbach G, Hartung M. Attempt to estimate the share of human Salmonella infections, which are attributable to Salmonella originating from swine [in German]. Berliner und Munchener Tierarztliche Wochenschrift. 1999;112:296-300.
8. Buzby JC, Jordan TR, MacDonald JM. Bacterial Foodborne Disease: Medical Costs and Productivity Losses. USDA. August 1996:1-81. Agriculture Economic Report No. (AER741).
9. Part III: Changes in the U.S. Pork Industry 1990-1995. USDA:APHIS:VS Centers for Epidemiology and Animal Health, 1997, N248.1097. Available at: http://www.aphis.usda.gov/vs/ceah/ncahs/nahms/swine/swine95/sw95Pt3.pdf . Accessed April 17, 2006.
10. Pathogen reduction; hazard analysis and critical control point (HACCP) systems; final rule. Federal Regist. 1996;61:38805-38855.
11. Salmonella Testing of Raw Meat and Poultry Products, 1998-2003. Progress Report. Available at: http://www.fsis.usda.gov/science/microbiology/index.asp . Accessed April 17, 2006.
12. Fedorka-Cray PJ, Dargatz DA, Thomas LA, Gray JT. Survey of Salmonella serotypes in feedlot cattle. J Food Prot. 1998;62:525-530.
13. Popoff MY, Le Minor L. Antigenic Formulas of the Salmonella Serovars. 8th ed. Paris: WHO Collaborating Centre for Reference and Research on Salmonella, Pasteur Institute; 2001.
*14. Bahnson PB, White B. Risk factors for the occurrence of drug resistant Salmonella spp. in commercial swine production. Proc Nat Pork Board Symp Antimicrob Resis Alternatives. West Des Moines, Iowa; 2004.
15. Carlson A, Blaha T. In-herd prevalence of Salmonella in 25 selected Minnesota swine farms. J Swine Health Prod. 2001;9(1):7-10.
16. Lammerding AM, Garcia MM, Mann ED, Robinson Y, Dorward WJ, Truscott RB, Tittiger F. Prevalence of Salmonella and thermophilic Campylobacter in fresh pork, beef, veal and poultry in Canada. J Food Prot. 1988;51:47-52.
17. Gebreyes WA, Davies PR, Turkson PK, Morrow MWE, Funk JA, Altiera C. Salmonella enterica serovars from pigs on farms and after slaughter and validity of using bacteriologic data to define herd Salmonella status. J Food Prot. 2004;67:691-697.
18. Hurd HS, McKean JD, Griffith RW, Wesley IV, Rostagno MH. Salmonella enterica infections in market swine with and without transport and holding. Appl Environ Microbiol. 2002;68:2376-2381.
19. Kasbohrer A, Protz D, Helmuth R, Nockler K, Blaha T, Conraths FJ, Geue L. Salmonella in slaughter pigs of German origin: an epidemiological study. Eur J Epidemiol. 2000;16:141-146.
20. Edel W, Kampelmacher EH. Epidemiological studies on Salmonella in a certain area ("Walcheren Project") II. Salmonella in the mesenteric lymph nodes and rectal contents of normal pigs. Zentralblatt fuer Bakteriologie Parasitenkunde Infektionskrankheiten und Hygiene Erste Abteilung Originale Reihe A Medizinische Mikrobiologie und Parasitologie. 1976;236:74-82.
21. Baggesen DL, Wegener HC, Bager F, Stege H, Christensen J. Herd prevalence of Salmonella enterica infections in Danish slaughter pigs determined by microbiological testing. Prev Vet Med. 1996;26:201-213.
22. Hogs and Pigs. December 28, 1999. National Agricultural Statistics Service (NASS), Agricultural Statistics Board, US Department of Agriculture. 1999. Available at: http://usda.mannlib.cornell.edu/reports/nassr/livestock/php-bb/1999/hgpg1299.pdf . Accessed April 24, 2006.
23. Lo Fo Wong DM, Dahl J, Wingstrand A, van der Wolf PJ, von Altrock A, Thorberg BM. European longitudinal study in Salmonella seronegative- and seropositive-classified finishing pig herds. Epidemiol Infect. 2004;132:903-914.
24. Funk JA, Davies PR, Nichols MA. Longitudinal study of Salmonella enterica in growing pigs reared in multiple-site swine production systems. Vet Microbiol. 2001;22:45-60.
25. Kranker S, Alban L, Boes J, Dahl J. Longitudinal study of Salmonella enterica serotype Typhimurium infection in three Danish farrow-to-finish swine herds. J Clin Microbiol. 2003;41:2282-2288.
26. Shedding of Salmonella by Finisher Hogs in the U.S. Info Sheet. United States Department of Agriculture, Animal and Plant Health Inspection Service, Veterinary Services, Centers for Epidemiology and Animal Health. Available at: http://www.aphis.usda.gov/vs/ceah/ncahs/nahms/swine/swine95/sw95salm.pdf . Accessed April 17, 2006.
27. Bahnson PB, Kim JY, Weigel RM, Miller GY, Troutt HF. Associations between on-farm and slaughter plant detection of Salmonella in market-weight pigs. J Food Prot. 2005;68:246-251.
28. FoodNet 1998 Annual Report. Centers for Disease Control and Prevention, March 2000. Available at: http://www.cdc.gov/foodnet/annual/98/FN_98_part_1.pdf . Accessed April 21, 2006.
29. Sarwari AR, Magder LS, Levine P, McNamara AM, Knower S, Armstrong GL, Etzel R, Hollingsworth J, Morris JG Jr. Serotype distribution of Salmonella isolates from food animals after slaughter differs from that of isolates found in humans. J Infect Dis. 2001;183:1295-1299.
30. Vugia DJ, Samuel M, Farley MM, Marcus R, Shiferaw B, Shallow S, Smith K, Angulo FJ. Invasive Salmonella infections in the United States, FoodNet, 1996 -1999: Incidence, serotype distribution, and outcome. Clin Infect Dis. 2004;38(Suppl 3):S149-S156.
31. Fedorka-Cray PJ, Kelley LC, Stabel TJ, Gray JT, Laufer JA. Alternate routes of invasion may affect pathogenesis of Salmonella typhimurium in swine. Infect Immun. 1995;63:2658-2664.
32. Hurd HS, Gailey JK, McKean JD, Rostagno MH. Rapid infection in market-weight swine following exposure to a Salmonella Typhimurium-contaminated environment. Am J Vet Res. 2001;62:1194-1197.
33. Janssen DW, Busta FF. Repair of injury in Salmonella anatum cells after freezing and thawing in milk. Cryobiology. 1973;10:386-392.
34. O’Carroll JM, Davies PR, Correa MT, Slenning BD. Effects of sample storage and delayed secondary enrichment on detection of Salmonella spp. in swine feces. Am J Vet Res. 1999;60:359-362.
35. Opara OO, Mallinson ET, Tate CR, Carr LE, Miller RG, Stewart L, Kelleher C, Johnston RW, Joseph SW. The effect of exposure, storage times, and types of holding media on the drag-swab monitoring technique for Salmonella. Avian Dis. 1992;36:63-68.
36. Jones BD, Falkow S. Salmonellosis: Host immune responses and bacterial virulence determinants. Ann Rev Immunol. 1996;14:533-561.
37. Roof MB, Kramer TT, Kunesh JP, Roth JA. In vivo isolation of Salmonella choleraesuis from porcine neutrophils. Am J Vet Res. 1992;53:1333-1336.
38. Dohoo I, Martin W, Stryhn H. Veterinary Epidemiologic Research. University of Prince Edward Island, Charlottetown, Prince Edward Island, Canada: AVC Inc; 2003.
39. Funk JA, Davies PR, Nichols MA. The effect of fecal sample weight on detection of Salmonella enterica in swine feces. J Vet Diagn Invest. 2000;12:412-418.
40. Bager F, Petersen J. Sensitivity and specificity of different methods for the isolation of Salmonella from pigs. Acta Vet Scand. 1991;32:473-481.
* Non-refereed references