Diagnostic notes |
Non refereed |
Defining swine influenza virus
Marie R. Gramer, DVM
Minnesota Veterinary Diagnostic Laboratory, Veterinary Population Medicine, College of Veterinary Medicine, University of Minnesota, 1333 Gortner Avenue, St Paul, MN 55108; Tel: 800-605-8787; Fax: 612-624-8707; E-mail: grame003@umn.ed
Cite as: Gramer MR. Defining swine influenza virus. J Swine Health Prod. 2005;13(3):157-160.
Also available as a PDF.
Swine influenza has evolved from a seasonal disease caused by a stable genotype of swine influenza virus (SIV) to a year-round, endemic respiratory disease caused by multiple SIV genotypes undergoing continuous change.1 This article will describe the structure of influenza viruses and provide information on how SIV isolates are defined by their antigenic and genetic characteristics.
Swine influenza virus structure
Influenza viruses are enveloped, segmented, single-stranded, negative-sense RNA viruses belonging to the family Orthomyxoviridae.2 The Orthomyxoviridae family consists of four genera – influenza A virus, influenza B virus, influenza C virus, and thogotovirus.2 As influenza A viruses cause disease in animals, birds, and humans,3 including swine, this genus will be discussed in detail.
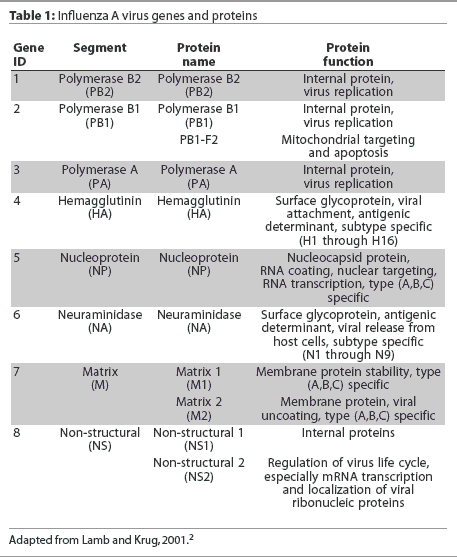
Eight RNA gene segments comprise the influenza A virus genome.2 These eight genes are responsible for the translation of 10 or 11 viral proteins.2,4,5 The highly conserved genes, nucleoprotein (NP) and matrix (M1), are used to classify influenza viruses as types A, B, and C. The hemagglutinin (HA) and neuraminidase (NA) genes encode for surface glycoproteins that project from the viral envelope. These glycoproteins possess distinct antigenic properties and nucleotide gene sequences and thus are used to subtype influenza viruses into 16 HA types (types 1 to 16) and nine NA types (types 1 to 9).2 Influenza A viruses are named by their HA and NA type, (eg, H1N1), and are often given "strain" names that include their genus or type, host species if other than human, location of isolation, arbitrary laboratory number, and year of isolation (eg, A/Swine/Iowa/15/1930). The influenza A virus gene segments, with the name of each gene, the protein encoded by it, and the function of the protein, are listed in Table 1.
Appearance of reassortant swine influenza viruses
For 80 years, only one strain of SIV, classical H1N1, was endemic in North America. Then, in 1998, a reassortant H3N2 SIV was detected in US swine.6 During 1998, reassortant H3N2 strains of SIV emerged in the swine population, that were either double reassortant, containing gene segments from human and avian strains of influenza (eg, A/Sw/NC/98) or triple reassortant, containing gene segments from human, avian, and swine influenza strains (eg, A/Sw/TX/98). In early 1999, classical H1N1 SIV reassorted with H3N2 SIV virus to create another reassortant virus, H1N2.7 Control was now further complicated, as was evidenced by multiple outbreaks of swine influenza caused by H1N2 infection in herds that had been vaccinated against classical H1N1 SIV.1,8 Further reassortment occurred in late 2002, wherein both the HA and NA genes of H3N2 SIV have been replaced by the classical H1 and N1 genes, thereby creating a reassortant, novel H1N1 SIV with avian internal genes (PA and PB2).9
These recently appearing influenza A viruses have complicated disease management and control, in large part because endemic SIV continually infects susceptible pigs,2 but also because several strains of SIV may be circulating in the herd concurrently.10 Some swine herds experience severe influenza-induced respiratory disease despite vaccination, with resulting poor growth performance and mortality. These performance and death losses are frustrating to producers and vaccine companies, both of which have requested that diagnostic laboratories and researchers perform detailed influenza characterization of SIVs isolated from pigs.
Virus characterization
Serotyping and antigenic characterization
Serological assays based on hemagglutination inhibition (HI) are historically the most common way to determine antigenic characteristics of influenza A viruses, and the HI test also serves as a functional assay for detection of anti-influenza antibodies in sera.11
The HI test exploits the ability of influenza viruses to agglutinate red blood cells (RBCs), a characteristic for which the HA gene was named. Thus, an influenza virus can be identified as to its subtype in an HI assay. A standard concentration of influenza virus (antigen) and serial dilutions of HA-subtype-specific antiserum are mixed, and after a short incubation period, RBCs are added to the antigen-antibody mixture.12 If the serum antibodies bind to the viral hemagglutinin, hemagglutination is inhibited, and the RBCs settle to the bottom of the test well. Hemagglutination reactions are compared among the sets of antisera and viral antigens in a test panel, and assumptions are made concerning the antigenic relatedness of the viruses on the basis of these HI reactions.12 For example, an HI reaction is observed in a well containing a 1:640 dilution of A/Swine/TX/1998 H3-specific antiserum and an unidentified SIV. Also in the test panel, there is an HI reaction in a well containing a 1:20 dilution of A/Swine/CO/1999 H3-specific antiserum with the unidentified virus, and no HI reaction in any of the wells containing serial dilutions of A/Sw/Iowa/1973 H1-specific antiserum. The homologous reaction between A/Sw/Texas/1998 H3-specific antiserum and A/Sw/Texas/1998 reference antigen occurs at the 1:1280 dilution. The interpretation of this HI assay results in the identification of the unidentified SIV as an H3 subtype, more specifically as an A/Sw/Texas/1998 H3-like variant of the H3 subtype, because the reaction is within a twofold dilution of homologous reaction and is fourfold greater than the reaction observed with the other antisera in the HI test panel.
Hemagglutination inhibition tests can also be used to characterize virus variants within a subtype. The above example demonstrated the ability of the HI test to discern different variants of H3 SIV when there are several reference sera in the panel. As a characterization or variant identification tool, the HI test is highly dependent on the number of reference antisera available.13 Reference antisera are usually generated by inoculation of ferrets, chickens, goats, or sheep with a specific subtype of influenza field virus for the production and collection of polyclonal antisera. For the production and collection of monospecific antisera, the animals or birds are inoculated with a purified HA fraction of influenza virus and postinoculation sera are then collected.12 Monoclonal antibodies may be generated using denatured fragments of HA, and the highly specific antibodies can then be used in rapid identification.14
Hyperimmunized-pig polyclonal antiserum used in HI tests is produced by inoculating pigs with the influenza virus of interest, boostering with a second inoculation several weeks later, and collecting the postinoculation antisera.15 In a study of the antigenic characteristics of SIV field isolates in the Netherlands, hyperimmunized or postinfection swine antisera used in HI panels appeared to crossreact with more isolates than did ferret antisera.16 Nevertheless, the apparent broad reactivity and lack of discrimination of hyperimmunized swine antisera can be overcome by increasing the number of different antisera used in the panel.17
Hemagglutination assays have several limitations. They require viable, cultivated virus, which may take several days to propagate.12 The virus, controls, and antisera must be properly diluted and titered at the beginning and end of every experiment.12 Repeatability of tests among experiments, technicians, and laboratories may interfere with comparisons of different experiments.17 The antisera used in the HI assays may not be able to distinguish antigenic differences between some viruses.17 Results of HI tests may also be disparate; for example, a heterologous reaction may have a higher titer than the homologous reaction.17 Finally, there may be a lack of symmetry between HI reactions; for example, reaction of serum 1 and antigen 1 may have a titer of 1:1280; reaction of serum 1 and antigen 2 may have a titer of 1:1280; but reaction of serum 2 and antigen 1 may have a titer of only 1:320.17 For these reasons, HI testing should be coupled with sequencing of the HA gene to correlate antigenic differences with genetic differences;18 with inoculation and challenge studies to compare in vivo results to the HI test results;19 or with mathematical and spatial analyses of antigenic relationships between viruses that may prove to be very informative.13
Genotyping and genetic characterization
Influenza virus genotyping is usually directed at the HA and NA genes,20 with additional information gained as needed from the internal genes, especially PA, PB2, and PB1.21 Reverse transcriptase-polymerase chain reaction (RT-PCR) may be used for genetic analysis of an RNA gene with known sequence, and RT-PCR tests are available for subtyping swine influenza viruses as H1N1, H1N2, H3N2, or H3N1. There are also RT-PCR tests for the internal genes, particularly PA and PB2. The advantages of PCR tests for HA and NA typing are that they are more specific and faster than serotyping, do not require viable virus or cultured virus, and can be automated. The disadvantages of using PCR for HA and NA typing include their requirements for skilled technicians and necessary equipment, and their inability to detect RNA if the HA or NA is significantly different than expected or has degraded beyond detection. Polymerase chain reaction tests will become more commonplace as they are validated and as unique variations of swine influenza virus, untypeable by serotyping, continue to occur.
Genetic sequence analyses of the HA and NA genes of circulating influenza viruses can reveal more than just the subtype. When studied carefully, they can reveal mutations in antigenic regions of the molecule that may have contributed to the virus’ ability to evade the immune system and spread.22 Sequence analysis of any of the genes can provide epidemiological information regarding origin of the virus and help to distinguish and identify subtype variants of different influenza viruses.23 Nucleotide sequencing, which determines the genetic code for the entire virus or a portion of it, is becoming more important as new and changing SIVs continue to emerge. The genetic makeup of the virus can be used to determine which viruses share a similar origin and are related to each other. Genetic analyses of the HA gene of the contemporary swine reassortant H1N1 viruses have revealed that these H1N1 subtype variants contain significant genetic differences that are apparently related to antigenic differences and escape from vaccinal immunity.
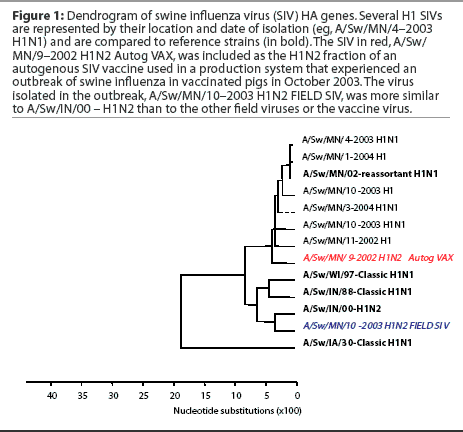
Subtyping and sequencing SIV isolates from disease outbreaks may provide useful information to the swine practitioner and producer, who may be seeking to alter vaccine strategies on the basis of the results. Figure 1 shows an example of a dendrogram or phylogenetic tree of swine influenza virus HA genes. In this dendrogram, several H1 SIVs are represented by their location and date of isolation (eg, A/Sw/MN/4-2003 H1N1) and are compared to reference strains (in bold). In red is the virus currently included as the H1N2 fraction of the autogenous SIV vaccine used in a production system that experienced an outbreak of swine influenza in vaccinated pigs in October 2003. This autogenous vaccine virus and all previous H1N1 and H1N2 isolates from this herd were of similar genotype and grouped with reference strain A/Sw/MN/02 – reassortant H1N1. Strain A/Sw/MN/02 – reassortant H1N1 is a contemporary SIV variant H1N1 that contains avian internal genes and differs in the HA gene from both the classical H1N1 and the prototypic H1N2 swine influenza virus isolated in Indiana in 2000 (A/Sw/IN/00 – H1N2). The virus isolated in the outbreak was subtyped as an H1N2, but genetic sequencing of the HA gene revealed that the outbreak strain (in blue) was more similar to A/Sw/IN/00 – H1N2 than to the other field viruses or to the vaccine virus. While timing of vaccination delivery may have also been an issue in this case, the presence of a genetically different virus was also examined as a contributing factor, and both the vaccine content and the timing issues were addressed.
 ,
,
Summary
While genetic analyses of influenza viruses have told us a great deal about the epidemiology and origin of SIVs in our swine population, they do not provide us with information as to whether SIVs with different genetic sequences will react the same way in the pig or whether disease will be prevented by administering the usual vaccine. Studies in Europe20,24 and in the United States25 have shown that even when antigenic (ie, serologic) and genetic differences exist among several circulating SIVs, vaccination with a single type of SIV vaccine may protect against the SIV variants if the pigs have sufficiently high concentrations of serum antibodies. It is important now, more than ever, to conduct cross-protection and challenge studies with the variant SIVs that are found in swine worldwide and to determine whether current vaccines will continue to be effective in our US swine population.
References
*1. Erickson GA, Gramer ML. North America perplexed by swine influenza. Pig Prog Resp Dis. October 2003:18-19.
2. Lamb RA, Krug RM. Orthomyxoviridae: The viruses and their replication. In: Fields BN, Knipe DM, Howley PM, eds. Fields Virology. 4th ed. Philadelphia, Pennsylvania: Lippincott-Raven; 2001:1487-1532.
3. Wright P, Webster R. Orthomyxoviruses. In: Fields BN, Knipe DM, Howley PM, eds. Fields Virology. 4th ed. Philadelphia, Pennsylvania: Lippincott-Raven; 2001:1533-1579.
4. Gibbs JS, Malide D, Hornung F, Bennink JR, Yewdell JW. The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J Virol. 2003;77:7214-7224.
5. Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O’Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. A novel influenza A virus mitochondrial protein that induces cell death. Nature Med. 2001;7:1306-1312.
6. Brown IH. The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol. 2000;74:29-46.
7. Karasin AI, Olsen CW, Anderson GA. Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana. J Clin Microbiol. 2000;38:2453-2456.
8. Karasin AI, Schutten MM, Cooper LA, Smith CB, Subbarao K, Anderson GA, Carman S, Olsen CW. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 2000;68:71-95.
9. Webby RJ, Rossow K, Erickson G, Sims Y, Webster R. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res. 2004;103:67-73.
10. Choi YK, Goyal SM, Kang SW, Fahrman MW, Joo HS. Detection and subtyping of swine influenza H1N1, H1N2 and H3N2 viruses in clinical samples using two multiplex RT-PCR assays. J Virol Methods. 2002;102:53-59.
11. Storch GA. Diagnostic virology. In: Fields BN, Knipe DM, Howley PM, eds. Fields Virology 4th ed. Philadelphia, Pennsylvania: Lippincott-Raven; 2001:493-531.
12. Manual on Animal Influenza Diagnosis and Surveillance. Geneva, Switzerland: World Health Organization; 2002. Available at: http://www.who.int/emc-documents/influenza/docs/animalinfluenza/html . Accessed February 15, 2005.
13. Smith DJ. Applications of bioinformatics and computational biology to influenza surveillance and vaccine strain selection. Vaccine. 2003;21:1758-1761.
14. Ziegler T, Hall H, Sanchez-Fauguier A, Gamble WC, Cox NJ. Type- and subtype-specific detection of influenza viruses in clinical specimens by rapid culture assay. J Clin Microbiol. 1995;33:318-321.
15. Richt JA, Lager KM, Janke BH, Woods RD, Webster RG, Webby RJ. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J Clin Microbiol. 2003;41:3198-3205.
16. de Jong JC, Heinen PP, Loeffen WLA, van Nieuwstadt AP, Claas ECJ, Bestebroer TM, Bijlsma K, Verweij C, Osterhaus ADME, Mimmelzwaan GF, Fouchier RAM, Kimman TG. Antigenic and molecular heterogeneity in recent swine influenza A (H1N1) virus isolates with possible implications for vaccination policy. Vaccine. 2001;19:4452-4464.
17. Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371-376.
18. Schweiger B, Zadow I, Heckler R. Antigenic drift and variability of influenza viruses. Med Microbiol Immunol. 2002;191:133-138.
19. Van Reeth K, Labarque G, De Clercq S, Pensaert M. Efficacy of vaccination of pigs with different H1N1 swine influenza viruses using a recent challenge strain and different parameters of protection. Vaccine. 2001;19:4479-4486.
20. Hungnes O. The role of genetic analysis in influenza virus surveillance and strain characterization. Vaccine. 2002;20:B45-B49.
21. Claas ECJ. Pandemic influenza is a zoonosis, as it requires introduction of avian-like gene segments in the human population. Vet Microbiol. 2000;74:133-139.
22. Abed Y, Hardy I, Li Y, Boivin G. Divergent evolution of hemagglutinin and neuraminidase genes in recent influenza A:H3N2 viruses isolated in Canada. J Med Virol. 2002;67:589-595.
23. Gubareva LV, Novikov DV, Hayden FG. Assessment of hemagglutinin sequence heterogeneity during influenza virus transmission in families. J Infect Dis. 2002;186:1575-1581.
24. Heinen PP, van Nieuwstadt AP, de Boer-Luijtze EA, Bianchi ATJ. Analysis of the quality of protection induced by a porcine influenza A vaccine to challenge with an H3N2 virus. Vet Immunol Immunopath. 2001;82:39-56.
*25. Gramer ML, Rossow KD. Epidemiology of swine influenza and implications of reassortment. Proc AD Leman Swine Conf. St Paul, Minnesota. 2004;69-73.
*Non-refereed references