Diagnostic notes |
Non refereed |
Improving rate of success in isolating Haemophilus parasuis from clinical samples
Simone Oliveira, DVM, MS, PhD
Research and Development, Newport Laboratories, 727 Oxford Street, Worthington, MN 56187; E-mail: oliv0107@umn.edu.
Cite as: Oliveira S. Improving rate of success in isolating Haemophilus parasuis from clinical samples. J Swine Health Prod. 2004;12(6):308-309.
Also available as a PDF.
Isolation of Haemophilus parasuis from clinical samples is still a challenge for many veterinarians. Although this organism may be detected in clinical samples by polymerase chain reaction (PCR),1 isolation is necessary for further characterization of isolates by serotyping2-4 and genotyping.5 Factors that may influence H parasuis isolation include selection of animals for sampling and handling of samples prior to submission to a diagnostic laboratory.6,7 This article will describe in detail some important measures that may improve the chances of isolating H parasuis from clinical samples.
Selection of animals for sampling
Haemophilus parasuis is rarely isolated from dead pigs. In order to improve chances of isolation, untreated pigs showing clinical signs characteristic of acute infection should be euthanized and sampled. Although pigs showing respiratory distress (abdominal breathing, coughing) and swollen joints are the best candidates for sampling, pigs with central nervous system signs may also be considered.6,7 It is important to differentiate acute from chronic lesions. Pigs that survive a nursery outbreak may develop clinical signs early in the finisher. These clinical signs may be associated with development of fibrosis in the thoracic and abdominal cavities. Isolation of H parasuis from chronically affected animals is usually unsuccessful.
Sampling sites
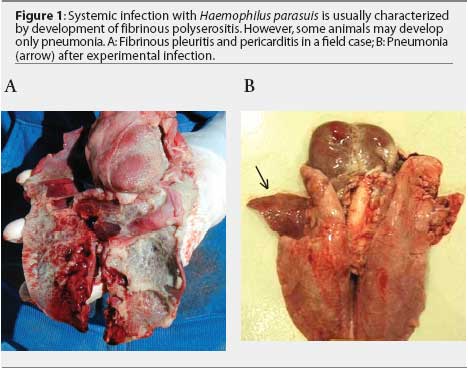
Non-respiratory sites are preferred for H parasuis isolation. Haemophilus parasuis is a commensal organism of the upper respiratory tract and may be isolated from the nasal cavity, tonsil, and trachea of healthy animals.8,9 Ideal sites for isolation are brain (meninges), pericardium, pleura, peritoneum, and joints. Lung tissue may be submitted when fibrinous pleuritis is observed (Figure 1A). In some cases, H parasuis induces only pneumonia (Figure 1B). Isolates recovered from pneumonic lungs may or may not represent the "problem" strain affecting the herd.5,7 Although H parasuis causes septicemia and is expected to be present in the blood during acute infection,10 isolation from blood samples collected from field cases is infrequent.
Sampling procedures
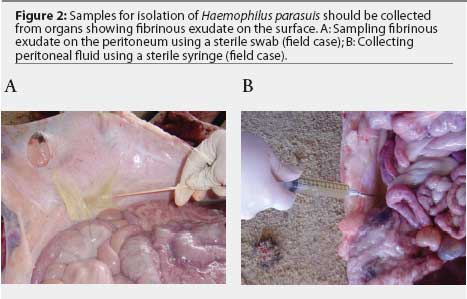
Samples for H parasuis isolation may be collected with sterile swabs placed in transport systems containing Stuart or Amies media. The Amies system maintains viability of H parasuis better than the Stuart system.11 Swabs should be collected from organs with fibrinous exudate on the surface (Figure 2A). Immunohistochemical studies have demonstrated that free H parasuis cells are usually concentrated in the fibrinous exudate.12 Fluids from joints, peritoneum, pericardium, and thoracic cavity may be collected using sterile syringes (Figure 2B). Haemophilus parasuis survives longer in whole tissues than in swabs. Because different H parasuis strains may be isolated from different body sites in the same pig (Figure 3), tissue samples should be submitted in separate bags. Live animals may be sampled without the need for euthanasia. Synovial fluid, for example, may be collected from swollen joints by using a sterile syringe and needle. Samples should be refrigerated immediately after collection.

Submission of samples for isolation
Haemophilus parasuis is temperature sensitive. Viable H parasuis organisms become undetectable in physiological saline at 42°C within 1 hour, at 37°C within 2 hours, and at 25°C within 8 hours.13 At refrigeration temperature (4°C), however, H parasuis can survive for a relatively long time.13,14 Samples collected for H parasuis isolation should be submitted to the diagnostic laboratory as soon as possible (1 to 2 days). Swabs, tissues samples, or syringes containing body fluids should be submitted in a Styrofoam container with ice packs. Tissues should be submitted fresh: the use of formalin is not necessary.
Diagnostic tests
Isolation of H parasuis is important for accurate characterization of the prevalent strains causing disease in the herd. After biochemical identification, H parasuis isolates may be further characterized by serotyping2-4 and genotyping5 (Figure 3). Both tests provide relevant information for disease control. Genotyping by means of enterobacterial repetitive intergenic consensus-PCR identifies the prevalent strains causing disease in affected herds.5 Euthanasia of 10 to 20 clinically affected nursery pigs allows for accurate identification of the prevalent H parasuis strains affecting the herd.14 When isolation attempts are unsuccessful, detection of H parasuis in clinical samples by PCR1 may be useful to define the role of this agent in mortality.
Summary
The rate of success in isolating H parasuis from clinical samples may be improved by following a few critical procedures. As this organism is very sensitive to high temperatures, samples should always be maintained under refrigeration. Transport systems containing Amies medium or submission of whole tissue tend to improve viability of H parasuis during transportation. It is important to collect samples from untreated, acutely infected pigs. Haemophilus parasuis is more likely to be isolated from pigs that have been euthanized than from dead pigs.
References
1. Oliveira S, Galina L, Pijoan C. Development of a PCR test to diagnose Haemophilus parasuis infections. J Vet Diagn Invest. 2001;13:495-501.
2. Kielstein P, Rapp-Gabrielson VJ. Designation of 15 serovars of Haemophilus parasuis on the basis of immunodiffusion using heat-stable antigen extracts. J Clin Microbiol. 1992;30:826-865.
3. del Río WL, Gutiérrez CB, Rodríguez Ferri EF. Value of indirect hemagglutination and coagglutination tests for serotyping Haemophilus parasuis. J Clin Microbiol. 2003;41:880-882.
4. Tadjine M, Mittal KR, Bourdon S, Gottschalk M. Development of a new serological test for serotyping Haemophilus parasuis isolates and determination of their prevalence in North America. J Clin Microbiol. 2004;42:839-840.
5. Oliveira S, Blackall PJ, Pijoan C. Characterization of the diversity of Haemophilus parasuis field isolates by serotyping and genotyping. Am J Vet Res. 2003;64:435-442.
*6. Oliveira S, Pijoan C, Morrison R. Role of Haemophilus parasuis in nursery mortality. Proc AD Leman Swine Conf. 2002:111-113.
7. Oliveira S, Pijoan C. Haemophilus parasuis: new trends on diagnosis, epidemiology and control. Vet Microbiol. 2004;99:1-12.
8. Vahle JL, Haynes JS, Andrews JJ. Interaction of Haemophilus parasuis with nasal and tracheal mucosa following intranasal inoculation of cesarean derived colostrum deprived (CDCD) swine. Can J Vet Res. 1997;61:200-206.
9. Moller K, Kilian M. V factor-dependent members of the family Pasteurellaceae in the porcine upper respiratory tract. J Clin Microbiol. 1990;28:2711-2716.
10. Amano H, Shibata M, Takahashi K, Sasaki Y. Effects on endotoxin pathogenicity in pigs with acute septicemia of Haemophilus parasuis infection. J Vet Med Sci. 1997;59:451-455.
11. del Rio ML, Gutiérrez B, Gutiérrez CB, Monter JL, Rodríguez Ferri EF. Evaluation of survival of Actinobacillus pleuropneumoniae and Haemophilus parasuis in four liquid media and two swab specimen transport systems. Am J Vet Res. 2003;64:1176-1180.
12. Segales J, Domingo M, Solano GI, Pijoan C. Immunohistochemical detection of Haemophilus parasuis serovar 5 in formalin-fixed, paraffin-embedded tissues of experimentally infected swine. J Vet Diagn Invest. 1997;9:237-243.
13. Morozumi T, Hiramune T. Effect of temperature on the survival of Haemophilus parasuis in physiological saline. Natl Inst Anim Health Q (Tokyo). 1982;22:90-91.
14. Oliveira S, Pijoan C, Morrison R. Evaluation of Haemophilus parasuis control in the nursery using vaccination and controlled exposure. J Swine Health Prod. 2004;12:123-128.
* Non-refereed reference.