Original research |
Peer reviewed |
Genetic diversity and possible avenues of dissemination of porcine reproductive and respiratory syndrome virus in two geographic regions of Mexico
Laura Batista, DVM, PhD; Carlos Pijoan, DVM, PhD; Humphrey Lwamba, BS; Craig R. Johnson, PhD; Michael P. Murtaugh, PhD
LB, CP: Department of Veterinary Population Medicine, University of Minnesota, St Paul, Minnesota. HL, CRJ, MPM: Department of Veterinary and Biomedical Sciences, University of Minnesota, St Paul, Minnesota. Corresponding author: Dr Laura Batista, Department of Clinical Sciences, University of Montreal, St-Hyacinthe, Québec, Canada J2S 7C6.
Cite as: Batista L, Pijoan C, Lwamba H, et al. Genetic diversity and possible avenues of dissemination of porcine reproductive and respiratory syndrome virus in two geographic regions of Mexico. J Swine Health Prod. 2004;12(4):170-175.
Also available as a PDF.
Summary
Objective: To determine the range of porcine reproductive and respiratory syndrome virus (PRRSV) genetic variation and possible avenues of dissemination in geographically separated swine herds in Mexico.
Methods: Pooled serum samples (n=33) from suspected viremic sows and nursery and finisher pigs from 13 sites in the state of Sonora (northwest Mexico) and six sites in Puebla (south-central region) were subjected to PRRSV polymerase chain reaction amplification, purification, and sequencing.
Results: Thirty-four PRRSV open reading frame 7 (ORF7) sequences were generated from a total of 31 isolates from 13 sites in Sonora and six in Puebla. The nucleotide similarity of ORF7 among isolates ranged from 86 to 100%. Phylogenetic analysis revealed no obvious geographic, temporal, or other characteristics that accounted for the observed distribution of isolates.
Implications: DNA sequence analysis revealed unexpectedly high levels of genetic variation among and within swine-rearing regions in Mexico, suggesting independent PRRSV introductions. Re-isolation of a vaccinal strain of PRRSV suggests importation of vaccinated animals or semen or surreptitious use. The diversity and distribution of viral genotypes indicates that the mechanisms of geographic spread were complex, such that effective control may present a substantial epidemiological challenge.
Keywords: swine, porcine
reproductive
and respiratory virus, epidemiology, sequencing
Search the AASV web site
for pages with similar keywords.
Received: April
14, 2003
Accepted: May
26, 2003
Porcine reproductive and respiratory syndrome virus (PRRSV) infection constitutes one of the major disease problems in the swine industry today.1,2 The causative agent is a small, enveloped, positive-strand RNA virus classified in the order Nidovirales, family Arteriviridae. Other family members include equine arteritis virus, lactate dehydrogenase-elevating virus, and simian hemorrhagic fever virus.3 These viruses replicate primarily in macrophages and are known to establish asymptomatic, persistent infections in their hosts. In an animal persistently infected with PRRSV, there is a continuous low level of viral replication, not a true steady-state persistent infection.4,5
Two distinct genotypes of PRRSV evolved independently in Europe and North America.6-8 Studies of PRRSV variation within North America have established that the virus is antigenically9 and genetically6,10 highly diverse. The major envelope glycoprotein gene, open reading frame 5 (ORF5), is most commonly used to analyze genetic variation.11-13 However, other regions of the viral genome that encode structural proteins, including open reading frame 7 (ORF7), which encodes the nucleocapsid (N) protein, demonstrate substantial genetic variation and have been used for genetic studies.6-8,10,13,14 Restriction fragment length polymorphism (RFLP) analysis also has been used to discriminate between vaccine and field isolates of PRRSV, but this method has not been adopted for genetic studies.15,16
We were interested in patterns of PRRSV genetic variation in the field, because differences in clinical signs and the apparent emergence of new syndromes may be due in part to changes in the PRRSV genome. This heterogeneity may pose a major obstacle for effective prevention and control of porcine reproductive and respiratory syndrome (PRRS). Therefore, the precise identification of PRRSV isolates by determination of nucleotide sequence of viral genetic material may be a valuable tool for understanding the success or failure of PRRS control and elimination procedures, possible sources of introduction into naive herds, and area spread. Here, we hypothesized that determination of genetic types of PRRSV isolates within a production system would allow a fuller understanding of the epidemiology of the disease. The objective of this study was to determine the range of PRRSV genetic variation and possible avenues of dissemination in geographically separated swine herds in Mexico.
Materials and methods
Serum samples
Pooled serum samples (n=33) were obtained from suspected viremic sows and nursery and finisher pigs from 13 herds in the state of Sonora in northwest Mexico and from six herds in Puebla in south-central Mexico. Five pigs from each production site were sampled, and sera were pooled to increase the possibility of finding a positive sample. Samples were submitted to a diagnostic laboratory, Investigacion Aplicada (Tehuacan, Puebla, Mexico), for testing by polymerase chain reaction (PCR) for PRRSV ORF7.17 Positive results were determined by agarose gel electrophoresis and ethidium bromide staining. The PCR products were spotted on 3MM paper (Whatman, Maidstone, England) and stored at 4°C until shipment to the University of Minnesota for reamplification and sequencing.
PCR reamplification
Samples of DNA were eluted from filters with 100 mL of water, purified (Qiagen PCR Purification kit; Valencia, California), and amplified with forward and reverse primers 5′- GTGGTAAACCTTGTCAAA TATGC-3′ and 5′-ATTCTTCCAATTCA AACACTG-3′ (nucleotides 2882-2904 and 3297-3317, respectively, of Genbank accession number U00153). Polymerase chain reactions were performed in a 50-ml volume containing 200 nM of each primer, 200 nM of each deoxynucleotide triphosphate, 1.25 units of Amplitaq DNA polymerase, 2 mM MgCl2 and 1x PCR buffer. All PCR reagents were obtained from Perkin-Elmer (Branchburg, New Jersey). Reactions were incubated for 10 minutes at 95°C, followed by 40 cycles of denaturation at 94°C for 30 seconds, annealing at 57°C for 30 seconds, and extension at 72°C for 45 seconds. A final extension was performed at 72°C for 7 minutes. The 436-bp PCR products were then visualized on 1% agarose gels stained with ethidium bromide.
Sequencing and sequence analysis
Products of PCR were extracted from the gel and purified (Qiagen Gel Extraction kit; Valencia, California) and either cloned into a plasmid vector (pGEM-T vector; Promega, Madison, Wisconsin) or submitted for sequencing to the Advanced Genetic Analysis Center, University of Minnesota (St Paul, Minnesota), where routine automated DNA sequencing was performed. Sequences were proofread manually, assembled, and analyzed phylogenetically with EDITSEQ, SEQMAN, and MEGALIGN programs of DNASTAR (Intelligenetics, Madison, Wisconsin), Clustal X18 and TreeView PPC.19 Clustal X and TreeView PPC are publicly available software programs.
Results
Thirty-one samples obtained from 13 sites in Sonora and six in Puebla were positive for PRRSV by PCR. Of the positive samples, 12 were from sows, 16 from nursery pigs, and three from finishers. A total of 34 PRRSV ORF7 sequences were generated from the 31 isolates. The sequencing trace files were unambiguous except for sample 29N, in which case the PCR product was cloned and ten individual clones were sequenced. The different nucleotide sequences were obtained, all of which produced the same amino acid sequence (Figure 1). The nucleotide sequences were deposited in Genbank with accession numbers AY209195 to AY209228.
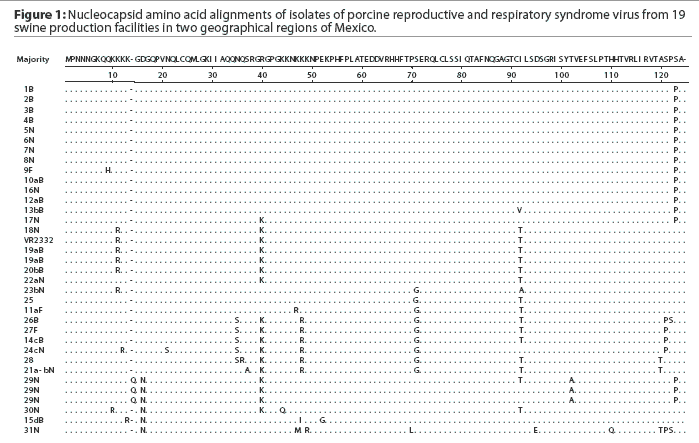
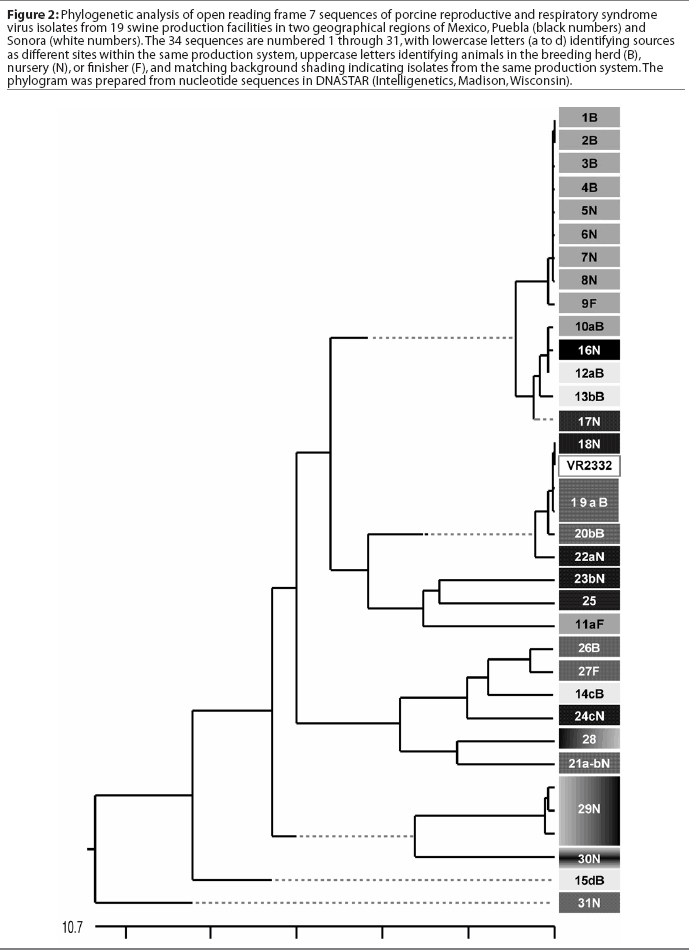
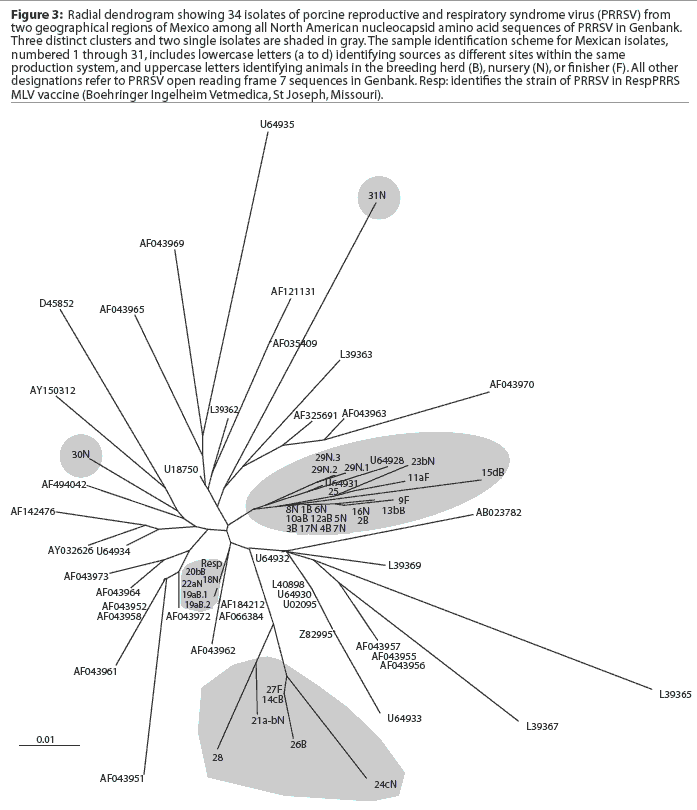
A comparison of the 34 sequences established that the nucleotide similarity of ORF7 among the isolates ranged from 86 to 100%. Nucleotide substitutions were located throughout the ORF (data not shown). Similarly, amino acid differences were numerous in all areas of the protein, as shown in the alignment in Figure 1. The maximum amino acid difference of 11.4% was observed between isolates 28 and 31N, and between isolates 24cN and 31N. A phylogram constructed from the 34 Sonora-Puebla nucleotide sequences and the type strain VR2332 is shown in Figure 2. Phylogenetic analysis revealed no obvious geographic, temporal, or other characteristics that accounted for the observed distribution of isolates. Figure 3, a radial dendrogram including all North American nucleocapsid amino acid sequences in Genbank, shows that all but two of the Mexican isolates clustered into one of three groups, shown in the shaded portions of the figure. One group consisted of apparent field re-isolates of Ingelvac PRRS MLV vaccine (previously marketed as RespPRRS; Boehringer Ingelheim Vetmedica, St Joseph, Missouri). A second group contained only isolates from this study and the nearest neighbor, Genbank U64932, which was isolated in Quebec, Canada, in 1993.20 Interestingly, the third and largest group of Mexican isolates contains two sequences, U64928 and U64931, which were isolated in Quebec, Canada, in 1991 and 1993, respectively.20 Isolates 30N and 31N had no close relatives.
As isolate 29N could not be resolved into a readable sequence upon initial sequencing, the 436-bp PCR product was cloned and 10 independent colonies were sequenced. Three distinct but closely related nucleotide sequences were identified. All 10 sequences contained a unique three-base insertion encoding the amino acid glutamine (Q) at position 14 (Figure 1). Thus, the protein contained 124 amino acids instead of the standard 123 amino acids reported in all other isolates except U64931, which has an asparagine (N) at position 43.
Discussion
The objective of this study was to describe the genetic variation of PRRSV field isolates recovered from sites in different geographical regions in order to understand the spread of the PRRSV in these two pork production areas in Mexico. A key element of the study was the use of diagnostic PCR samples for genetic analysis. Many PRRSV diagnostic PCR assays amplify ORF7, whereas genetic studies tend to be focused on ORF5. Here, genetic analysis of ORF7 revealed diverse PRRSV isolates in Mexico. Thus, ORF7 displays substantial genetic variation of epidemiological relevance. Use of diagnostic PCR also facilitates genetic analysis of field isolates, as a diagnostic PCR sample isolated locally may be shipped to centralized sequencing facilities for genetic analysis, internationally if necessary.
The reamplification performed at the University of Minnesota was necessary because the samples recovered from filter paper were insufficient for high quality sequencing. Direct sequencing of PCR products from pooled samples without subcloning, as performed in this study, has the risk of missing minor subpopulations of PRRSV isolates. However, pigs appear to usually harbor a single PRRSV genotype, even when directly inoculated with multiple strains;21,22 the trace files from sequencing are usually unambiguous; and the subcloning and sequencing that were performed in this study revealed closely related forms in only one isolate (29N).
The phylogram in Figure 2 reveals a high level of genetic variability in the ORF7 gene in the isolates from Sonora and Puebla, showing 10 different clusters (1B-9F, 10aB-17N, 18N-22aN, 23bN-11aF, 26B-24cN, 28-21a-bN, 29N, 30N, 15dB, and 31N). Isolates 15dB and 31N were very different from the other isolates, suggesting that they evolved from a very distinct ancestor. Isolates recovered from experimentally infected pigs tend to have an average of 0.1 to 0.2% change or less in ORF7, compared to the original isolate used to infect the pig.23 This rate stayed the same or declined with continued passage of recovered virus through additional pigs.23 Isolates with few differences are interpreted as being closely related, whereas isolates with substantial differences are interpreted as being independent.13 Since this was the first phylogenetic analysis of PRRSV in Mexico, we do not know if the variability noted in these results is characteristic of isolates from other areas of Mexican swine production. However, another phylogenetic analysis in the midwestern United States revealed a large amount of variability among local as well as regional PRRSV isolates.12 Also, in that study, phylogenetic analysis indicated that PRRSV isolates did not generally sort into location-specific groups. This observation agrees with our finding that genotypic variation among isolates did not correlate well with geographical proximity or, in some cases, with source of semen, breeding stock replacement, or both. Interestingly, in several instances, isolates from the two geographical regions, Sonora and Puebla, appeared to be closely related, even though the production sites of origin were separated by several thousand kilometers. Since we could identify no factor in common between these two areas, it implied the possibility of a common infectious source at the time the sites were stocked approximately 5 years previously, with limited genetic change thereafter. The opportunity for a more recent common source of infection could not be identified, but cannot be completely excluded. Additionally, isolates 12aB and 13bB, and 19aB and 20bB, from pigs in two different breeding herds in the same production system, were very similar, suggesting either little genetic change through the years or an inapparent means of virus spread. Isolates 1B, 2B, 3B, 4B, 5N, 6N, 7N, and 8N were 100% identical and 99% similar to isolate 9F. However, there was at least a 10% difference between strains 10aB and 11aF, even though they came from different sites in the same system. This difference might be due to horizontal transmission from nearby swine facilities that were not part of the production systems in this study. When isolates in the cluster containing isolates 12aB, 13bB, and 10aB are compared, a common source of infection for sites in different production systems located in the same geographical area appears to be likely. Isolates 12aB and 13bB originated from the same system, whereas isolate 10aB originated from a different system 24 km distant, with no common source of transport, feed, semen, or any other material. Isolates from two breeding herds (isolates 19aB and 20bB) and from their single combined nursery (isolate 21a-bN) were 100% identical to the Ingelvac MLV vaccine strain. This finding was unexpected, since there is no record of use of any type of PRRS vaccine, and the Ingelvac MLV was not licensed in Mexico. We hypothesize that the site acquired the vaccine strain directly or indirectly via vaccinated breeding stock or semen of vaccinated boars, or that there was surreptitious use of the vaccine. It is unlikely that field strains homologous to vaccine were re-isolated, since the vaccine was derived from a Minnesota field strain isolated in 1989 and never unequivocally isolated again. These unexpected results are also unlikely to have been caused by laboratory contamination, since neither Ingelvac MLV nor VR2332 strains were in use in either laboratory in Mexico or the United States when these studies were being performed.
Breeding herd isolates had only a 90% similarity with nursery isolates. Isolates 22aN, 23bN, and 22cN, which showed 90 to 96% similarity, were from three different sites in the same production system. The substantial variation among isolates, as shown in Figure 3, indicated that PRRSV introductions occurred repeatedly in Sonora and Puebla. Finally, strain 29N contained a novel insertion of three base pairs (CAA), encoding the extra amino acid glutamine at position 14. The emergence of this novel genotype may have occurred in response to immunological selection of mutants that escaped neutralizing antibodies or cytotoxic T lymphocytes, or as a result of defective viral particle production.4,24,25 It will be interesting to determine if this strain has different neutralization or T-cell reactivity characteristics consistent with immune escape.
Genetic analysis of PRRSV and clinical observations of the disease suggest that the virus is changing rapidly. The mechanisms of change involve mutation, genetic recombination, and horizontal gene transfer.10 The data from Mexico are consistent with the concept of rapid change, since two genetic clusters, both of which showed phylogenetic similarities to independent isolates from 1993 in Quebec, Canada, displayed substantial differences in variation within each cluster, and one strain, 29N, contained an insertion.
Characterization of the variation in PRRSV sequences is useful because it allows reconstruction of patterns of virus change in the field. It also allows one to determine if virus strains recovered from farms originate from pre-existing isolates or represent new introductions from an outside source. This method is useful in investigating differences and similarities among isolates in a group of farms having similar potential contamination sources, including semen, breeding stock, personnel, and business vehicles, and in hypothesizing about area spread and routes of transmission in a given geographical area. It is also an important tool to adjust acclimatization protocols that use gilt exposure to homologous strains within a farm.26
As of today, sequence information cannot be used to make inferences about biological properties of PRRSV. Therefore, questions regarding the impact of PRRSV diversity on the severity of clinical signs and manifestations, persistence, and immunity remain to be answered if we hope to successfully prevent, control, and eventually permanently eradicate PRRSV from swine production.
Implications
- High levels of genetic variation among and within two swine-producing regions in Mexico suggest that independent PRRSV introductions occurred.
- Vaccine re-isolation suggests that the vaccine strain was acquired directly or indirectly via vaccinated breeding stock or semen of vaccinated boars, or that there was surreptitious use of the vaccine.
- The diversity and distribution of viral genotypes indicate that the mechanisms of geographic spread were complex, such that effective control may present a substantial epidemiological challenge.
References
*1. Loula TJ. Mystery disease. Agri Prac. 1991;12:23-34.
2. Dee SA, Joo HS, Polson DD, Marsh WE. Evaluation of the effects of nursery depopulation of the profitability of 34 pig farms. Vet Rec. 1997;140:498-500.
3. Plagemann PGW, Moennig V. Lactate dehydrogenase-elevating virus, equine arteritis virus, and simian hemorrhagic fever virus: a new group of positive-strand RNA viruses. Adv Virus Res. 1992;41:99-192.
4. Domingo E, Baranowski E, Ruiz-Jarabo CM, Martin-Hernandez AM, Saiz JC, Escarmis C. Quasispecies structure and persistence of RNA viruses. Emerg Infect Dis. 1998;4:521-527.
5. Allende R, Laegreid WW, Kutish GF, Galeota JA, Wills RW, Osorio FA. Porcine reproductive and respiratory syndrome virus: description of persistence in individual pigs upon experimental infection. J Virol. 2000;74:10834-10837.
6. Meng XJ, Paul PS, Halbur PG, Lum MA. Phylogenetic analyses of the putative M (ORF6) and N (ORF7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the USA and Europe. Arch Virol. 1995;140:745-755.
7. Murtaugh MP, Elam MR, Kakach LT. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus. Arch Virol. 1995;140:1451-1460.
8. Nelsen CJ, Murtaugh MP, Faaberg KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. 1999;73:270-280.
9. Yoon KJ, Wu LL, Zimmerman JJ, Platt KB. Field isolates of porcine reproductive and respiratory syndrome virus (PRRSV) vary in their susceptibility to antibody dependent enhancement (ADE) of infection. Vet Microbiol. 1997;55:277-287.
10. Kapur V, Elam MR, Pawlovich TM, Murtaugh MP. Genetic variation in porcine reproductive and respiratory syndrome virus isolates in the midwestern United States. J Gen Virol. 1996;77:1271-1276.
11. Andreyev VG, Wesley RD, Mengeling WL, Vorwald AC, Lager KM. Genetic variation and phylogenetic relationships of 22 porcine reproductive and respiratory syndrome virus (PRRSV) field strains based on sequence analysis of open reading frame 5. Arch Virol. 1997;142:993-1001.
12. Goldberg TL, Hahn EC, Weigel RM, Scherba G. Genetic, geographical and temporal variation of porcine reproductive and respiratory syndrome virus in Illinois. J Gen Virol. 2000;81:171-179.
*13. Murtaugh MP, Faaberg K. How to interpret and use PRRSV sequence information. Proc A.D. Leman Swine Conf. 2001;28:60-66. Available on AASV CD-ROM
*14. Murtaugh MP, Faaberg KS, Laber J, Elam MR, Kapur V. Genetic variation in the PRRS virus. Adv Exper Med Biol. 1998;440:787-794.
15. Wesley RD, Mengeling WL, Lager KM, Clouser DF, Landgraf JG, Frey ML. Differentiation of a porcine reproductive and respiratory syndrome virus vaccine strain from North American field strains by restriction fragment length polymorphism analysis of ORF5. J Vet Diagn Invest. 1998;10:140-144.
16. Wesley RD, Mengeling WL, Lager KM, Vorwald AC, Roof MB. Evidence for divergence of restriction fragment length polymorphism patterns following in vivo replication of porcine reproductive and respiratory syndrome virus. Am J Vet Res. 1999;60:463-467.
17. Christopher-Hennings J, Nelson EA, Nelson JK, Hines RJ, Swenson SL, Hill HT, Zimmerman JJ, Katz JB, Yaeger MJ, Chase CC, Benfield DA. Detection of porcine reproductive and respiratory syndrome virus in boar semen by PCR. J Clin Microbiol. 1995;33:1730-1734.
18. Thompson JD, Gibson TJ, Plweniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;24:4876-4882.
*19. Page RDM. TreeView software. Available at http://taxonomy.zoology.gla.ac.uk/rod/rod.html. Accessed April 15, 2004.
20. Dea S, Gagnon CA, Mardassi H, Milane G. Antigenic variability among North American and European strains of porcine reproductive and respiratory syndrome virus as defined by monoclonal antibodies to the matrix protein. J Clin Microbiol. 1996;34:1488-1493.
21. Mengeling WL, Lager KM, Vorwald AC, Koehler KJ. Strain specificity of the immune response of pigs following vaccination with various strains of porcine reproductive and respiratory syndrome virus. Vet Microbiol. 2003;93:13-24.
22. Lager KM, Mengeling WL, Wesley RD. Strain predominance following exposure of vaccinated and naive pregnant gilts to multiple strains of porcine reproductive and respiratory syndrome virus. Can J Vet Res. 2003;67:121-127.
23. Chang CC, Yoon KJ, Zimmerman JJ, Harmon KM, Dixon PM, Dvorak CMT, Murtaugh MP. Evolution of porcine reproductive and respiratory syndrome (PRRS) virus during sequential passages in pigs. J Virol. 2002;76:4750-4763.
24. Duarte EA, Novella I, Weaver SC, Domingo E, Wain-Hobson S, Clarke DK, Moya A, Elena SF, de la Torre JC, Holland JJ. RNA virus quasispecies: significance for viral disease and epidemiology. Infect Agents Dis. 1994;3:201-214.
25 Ahmed R, Morrison LA, Knipe DM. Persistence of viruses. In: Fields BN, Knipe DM, Howley PM, eds. Field’s Virology. Philadelphia: Lippencott-Raven; 1996:219-249.
26. Batista L, Pijoan C, Torremorell M. Experimental exposure to porcine reproductive and respiratory syndrome virus (PRRSV) in gilts during acclimatization. J Swine Health Prod. 2002;10:147-150.
* Non-refereed references.