Original research |
Peer reviewed |
Genotypic and phenotypic comparison of swine Salmonella isolates from farm and abattoir
Matthew M. Erdman, BS; Stephanie D. Wedel, BS; D.L. Harris, DVM, PhD
MME: Department of Microbiology, College of Agriculture, 207 Science 1, and Department of Veterinary Microbiology, College of Veterinary Medicine, Iowa State University, Ames, IA 50011. SDW: Department of Microbiology, College of Agriculture, 207 Science 1, Iowa State University, Ames, IA 50011. DLH: Department of Microbiology, College of Agriculture, 207 Science 1, and Department of Veterinary Diagnostic and Production Animal Medicine, College of Veterinary Medicine, Iowa State University, Ames, IA 50011.Corresponding author: Dr D.L. (Hank) Harris, Department of Microbiology, 207 Science 1, Iowa State University, Ames, IA 50011; Tel: 515-294-1664; Fax: 515-294-6019; E-mail: hharris@iastate.edu.
Erdman MM, Wedel SD, Harris DL. Genotypic and phenotypic comparison of swine Salmonella isolates from farm and abattoir. J Swine Health Prod. 2003;11(4):169-172. Also available as a PDF.
Summary
Objectives: To determine if pigs became infected with different serotypes of Salmonella following transport and lairage, and whether genotypic and phenotypic analysis would identify different subtypes within the same serotypes.
Methods: Salmonella organisms were isolated from lymph nodes of pigs from three herds (A,B,C) at the abattoir, and isolates were compared to those previously cultured from fecal samples from two of the herds (A,B). The farms were located in three different states within the United States, and pigs were transported to the same abattoir in separate clean and disinfected vehicles. Serotyping, antibiotic resistance profiling, and genetic fingerprinting using pulsed- field gel electrophoresis were used to compare isolates.
Results: Salmonella serovars recovered from feces of pigs from farms A and B were also recovered from lymph nodes of pigs from the same farm. Additionally, isolates cultured from pigs at the abattoir were distinguishable from those cultured from pigs on the source farm by identification of serovars, genetic fingerprinting within serovars, and antibiotic resistance profiles.
Implications: Biosecurity, from the time market pigs leave the farm up to and including the hours immediately prior to slaughter, is crucial to the control of salmonellae in pork. New populations of salmonellae, not previously present on the source farm, may be recovered from pigs at the abattoir. Rapid infection may occur in pigs not previously harboring Salmonella organisms, and the strain may be multi-antibiotic resistant, further elevating the food safety risk.
Keywords: swine, Salmonella, food safety, pulsed-field gel electrophoresis, antibiotic resistance Search for similar articles on this site
Received: July 10, 2002
Accepted: November 18, 2002
Understanding the epidemiology of human foodborne pathogen transmission through the "farm to fork" continuum is crucial when considering food safety in the livestock industry. In 1987, Morgan et al1 described the effect of time in lairage on Salmonella contamination of slaughter pigs. They showed that the percentage of pigs in which salmonella organisms could be cultured from the cecum increased directly with the time spent in lairage. It was suggested that both the size of the holding pen and its hygiene contributed to this result. In turn, a high isolation rate from the cecum was generally associated with Salmonella recovery from the carcass.1
In 1995, Fedorka-Cray et al2 described an alternate route of invasion in which Salmonella serovar Typhimurium was isolated from lymph nodes and cecum 3 hours after intranasal inoculation of esophagotomized pigs. Recently, Salmonella infection has been shown to occur in pigs less than 2 to 3 hours after they were placed in pens which previously housed Salmonella infected pigs.3 In addition, a greater prevalence of Salmonella serovars, including serovars not recovered from cohort pigs at the farm of origin, have been reported in pigs at slaughter.4 Recent work by others suggests that acute infection might play a role in contamination of pigs at the slaughterhouse.5,6 Thus, it appears possible that, from the time they leave the farm until they are slaughtered, pigs from Salmonella-free herds may become contaminated through direct contact with contaminated trucks or facilities, or by commingling with Salmonella-positive pigs.4,7
In this three-herd study, we confirmed the previous work regarding new serovars being acquired in lairage. Our objective was to determine if new clones within serovars, distinguishable by genetic analysis and antibiotic profiling, were being acquired in lairage as well.
Materials and Methods
Sample collection
From July 1999 to July 2000, pooled pen fecal (PPF) samples were cultured approximately every other month from Herd A located in Kentucky and from Herd B located in Oklahoma. On each occasion, 20 PPF samples were collected, each consisting of 5 g of feces from five different places within a pen, for a total of 25 g. The sampled pens were chosen randomly and contained approximately 25 pigs weighing nearly 110 kg each. The samples were placed on ice and shipped overnight for culture within 24 hours of collection. In addition, during the course of this study, pigs that were sick or died were routinely necropsied, and tissues were submitted to a diagnostic laboratory for culture.
Approximately monthly from January 2000 to May 2000, 50 to 100 ileocecal lymph nodes were collected at an abattoir in Missouri from market weight pigs of Herds A and B, weighing approximately 120 kg and identified by tattoo. Pigs were transported from their respective farms to the same abattoir via separate clean and disinfected vehicles. Pigs used in the study were the first to be slaughtered that day and were held in lairage for at least 3 hours. Lymph nodes were also collected from pigs designated "Herd C," originating in Missouri and representing all other pigs killed that day. After Herd A, B, and C carcasses had been eviscerated, approximately 5 g of ileocecal lymph node was collected from each pig (n=359) and placed on dry ice to be cultured within 12 hours.
Culture
Pooled pen fecal samples were diluted 1:10 in buffered peptone water (BPW; Difco, Detroit, Michigan). Lymph nodes were dipped in 70% ethanol for 2 seconds, flamed to decontaminate, macerated, and diluted 1:10 in BPW. Both fecal and lymph node BPW suspensions were incubated for 24 hours at 37°C, then 0.1-mL samples of the BPW suspensions were transferred for selective enrichment in Rappaport-Vassiladis (RV) broth (Difco), which was incubated at 42°C for 24 hours. A 0.1-mL sample of each BPW suspension was also plated on selective media, xylose lysine deoxycholate agar (XLD), at 37°C for 24 hours. A maximum of five Salmonella suspect colonies per XLD plate were inoculated into tubed media (Kliglers, sulfide indole motility, phenylalanine, and lysine iron tubes) and were tested for agglutination with Salmonella O antisera (Difco).8 All Salmonella isolates were serotyped at the National Veterinary Service Laboratories, Ames, Iowa.
Antimicrobial sensitivity testing
Antibiograms were determined using the Kirby-Baur disk diffusion method.9 Salmonella isolates were suspended in Mueller-Hinton (MH) broth (Difco), and turbidity was adjusted to 0.5 MacFarland Standard. Cultures were then inoculated onto MH plates and antimicrobial discs were applied (BBL, Cockeysville, Maryland). Plates were incubated at 37°C for 24 hours, and the zone of inhibition was interpreted according to the manufacturer’s instruction. Isolates were checked for resistance to the following antibiotics: amoxicillin-clavulanic acid, ampicillin, aztreonam, cefepime, cefotaxime, cefoxitin, ceftazidime, ceftiofur, ceftriaxone, cephalothin, chloramphenicol, ciprofloxacin, colistin, enrofloxacin, furazolidone, gentamicin, kanamycin, naladixic acid, neomycin, piperacillin, piperacillin-tazobactam, spectinomycin, streptomycin, sulfisoxazole, tetracycline, ticarcillin-clavulanic acid, trimethoprim, and trimethoprim-sulfamethoxazole.
Genotypic analysis
Pulsed-field gel electrophoresis (PFGE) of Salmonella serovar Heidelberg isolates (n=68) was conducted using the endonuclease XbaI according to the protocol utilized by the Centers for Disease Control and Prevention.10 The PFGE patterns were analyzed visually and also compared using BioNumerics software (Applied Maths, Kortrijk, Belgium). Dendrograms were constructed by the unweighted pair group method using arithmetic averages, Dice coefficient, and 0.8% optimization with 1.0% band position tolerance.11
Results
Salmonella serovars present in PPF samples from Herds A and B were also present within lymph nodes of pigs from the respective herds at slaughter (Table 1). Additional serovars and different subtypes within serovars were also isolated from lymph nodes of pigs from Herds A and B. Serovars that had not been isolated from PPF samples on the farm during the time frame of this study were isolated from pigs of Herd B (Table 1). In addition, serovars with antibiotic profiles not previously identified in the herd were found in lymph nodes at slaughter (Table 2). In both Herds A and B, at least one Salmonella subtype isolated from lymph nodes was resistant to more antibiotics than any subtype isolated from the respective herd PPF samples.
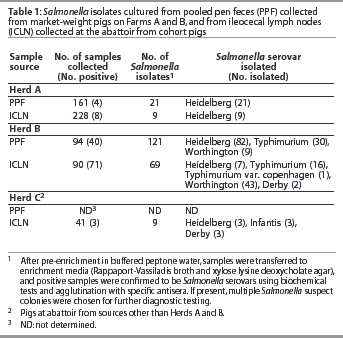
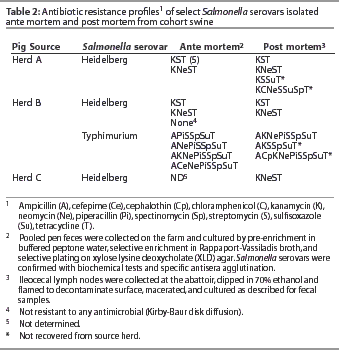
Salmonella Heidelberg was present in PPF samples of pigs from both Herds A and B and also from the lymph nodes of pigs from Herds A, B, and C, and was thus further analyzed by PFGE (Figure 1). A single PFGE pattern represented all of the Salmonella Heidelberg isolates from Herd A PPF samples. While this same pattern occurred in isolates from Herd A lymph nodes, isolates with a unique PFGE pattern were also present that were only 95% related. A single PFGE pattern also represented all Salmonella Heidelberg isolates from the Herd B PPF samples, although this pattern was slightly different from that of the Herd A PPF samples. This pattern was also observed in the isolate from the Herd B lymph nodes, but again a second pattern occurred. This new pattern was less than 80% related to the isolate from Herd B, but was exactly the same as the PFGE pattern of Salmonella Heidelberg isolates from lymph nodes of pigs from Herd C. No relationship was seen between serovar antibiotic profiles and PFGE patterns.
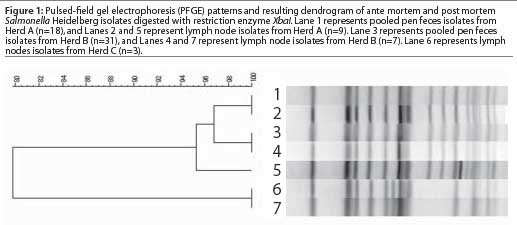
Salmonella was isolated from the lymph nodes submitted to a diagnostic laboratory from two Herd A pigs from that had been necropsied on the farm. The isolates were Salmonella Heidelberg and had PFGE patterns identical to the on-farm fecal isolates (data not shown).
Discussion
We found, in pigs at slaughter, Salmonella serovars and subtypes of serovars (distinguishable either by molecular genetic analysis or antibiotic resistance profiles) that were different from serovars found in the feces of cohort pigs at the farms of origin. The Salmonella Heidelberg isolates from farm and abattoir originating from the same source, while closely related, were still distinguishable by PFGE profiling. Previous work has shown a significant increase in Salmonella prevalence in lymph nodes of pigs slaughtered at an abattoir compared to those slaughtered on the farm from the same cohort.4 The isolation of different Salmonella serovars from the same cohort of pigs at the abattoir supports previous work showing an increase in serodiversity following transport and lairage.4 Between the time when the pigs left the farm and when they were slaughtered, they became infected with an additional Salmonella serovar, and likely new subtypes within serovars, that rapidly disseminated to the ileocecal lymph nodes.
This study assumed that the Salmonella serovars and genotypes detectable in feces are representative of the isolates present in lymph nodes in pigs at the herd of origin. This is a reasonable assumption based on the work of Wood et al,12 who found that the strain of Salmonella Typhimurium used to inoculate 7- to 8-week-old pigs was consistentlycultured from the feces as well as from the lymph nodes of pigs necropsied up to 28 weeks post-exposure. A recent study did indicate that no Salmonella, or fewer serovars, were present in feces compared to lymph nodes when pigs were necropsied on the farm.4 However, in that study, only 1-gram samples of feces were collected from individual pigs, rather than pooled pen fecal samples. Funk et al13 showed that a 25-gram sample of feces is far superior to a 1-gram sample for Salmonella detection. In addition, when pigs were necropsied on the farm and samples were subsequently submitted to a diagnostic laboratory for culture, Salmonella isolates from the lymph nodes were the same as those from PPF samples of pigs in the same cohort.
Efforts to reduce Salmonella rely on establishment of intervention strategies. The results of this study confirm the work of others,2-4 ie, we conclude that acute Salmonella infection during transport and lairage requires intervention in order to control Salmonella contamination of pork. If indeed pigs exposed to Salmonella hours before slaughter can become systemically infected, pigs from Salmonella-free sources, or sources with a low prevalence of salmonellae,14 may pose the same threat to food hygiene as pigs from highly contaminated systems. In addition, pork may become contaminated with organisms that are resistant to more antibiotics, thus posing a further threat to human health.
Implications
- Salmonella serovars and subtypes within serovars not previously identified on the farm of origin can be recovered from pigs at the abattoir.
- Pigs from herds with a low prevalence of Salmonella that come in contact with contaminated trucks, facilities, or pigs may pose the same risk to food safety as those from herds with a high prevalence of Salmonella.
- The time immediately prior to slaughter, including both transport and lairage, should be considered a key intervention point for control of Salmonella in pork production.
Acknowledgements
This work was supported by PIC, USA. We would also like to thank Jeanne Nugent, Brad Chriswell, Stephen Gaul, Carrie Basak, Alan Loynachan, Daron Allen, Justin Bruch, and Jeff Forsyth for technical assistance.
References – refereed
1. Morgan IR, Krautil FL, Craven JA. Effect of time in lairage on caecal and carcass Salmonella contamination of slaughter pigs. Epidemiol Infect. 1987;98:323-330.
2. Fedorka-Cray PJ, Kelley LC, Stabel TJ, Gray JT, Laufer JA. Alternate routes of invasion may affect pathogenesis of Salmonella typhimurium in swine. Infect Immun. 1995;63:2658-2664.
3. Hurd HS, Gailey JK, McKean JD, Rostagno MH. Rapid infection in market-weight swine following exposure to a Salmonella Typhimurium-contaminated environment. Am J Vet Res. 2001;62:1194-1197.
4. Hurd HS, McKean JD, Griffith RW, Wesley IV, Rostagno MH. Salmonella enterica infections in market swine with and without transport and holding. Appl Envir Microbiol. 2002;68:2376-2381.
9. Performance standards for antimicrobial disk susceptibility tests for bacteria that grow aerobically. Villanova, Pennsylvania: National Committee for Clinical Laboratory Standards; 1997.
10. Bender JB, Hedberg CW, Boxrud DJ, Besser JM, Wicklund JH, Smith KE, Osterholm MT. Use of molecular subtyping in surveillance for Salmonella enterica serotype Typhimurium. N Eng J Med. 2001;344:189-195.
11. Markogiannakis A, Tassios PT, Lambiri M, Ward LR, Kourea-Kremastinou J, Legakis J, Vatopoulis AC. Multiple clones within multidrug-resistant Salmonella enterica serotype typhimurium phage type DT104. The Greek Nontyphoidal Salmonella Study Group. J Clin Microbiol. 2000;38:1269-1271.
12. Wood RL, Pospischil A, Rose R. Distribution of persistent Salmonella typhimurium infection in internal organs of swine. Am J Vet Res. 1989;50:1015-1021.
13. Funk J, Davies PR, Nichols MA. The effect of sample weight on detection of Salmonella enterica in swine feces. J Vet Diagn Invest. 2000;12:412-418.
14. Alban L, Stege H, Dahl J. The new classification system for slaughter-pig herds in the Danish Salmonella surveillance-and-control program. Prev Vet Med. 2002;53:133-146.
References – non refereed
5. Blaha TH, Solano-Aguilar G, Pijoan C. The early colonization pattern of S. Typhimurium in pigs after oral intake. Proc 2nd Int Sym Salm Pork 1997:71-73.
6. Nugent JM, Erdman M, Harris DL. Dissemination of Salmonella choleraesuis 54 and Salmonella choleraesuis 38 in swine [abstract]. Proc Conf Res Work Anim Dis. 2001.
7. Quessy S, Letellier A. Detection of animals and herds positive to Salmonella. Proc West Can Assoc Swine Pract. 2000:93-95.
8. Erdman M, Torremorell M, Harris IT, Harris DL. Monitoring Salmonella levels in pre-slaughter pigs using serology and pen feces culture. Proc AASP. Indianapolis, Indiana. 2000;163-165.