Diagnostic notes |
Non refereed |
Diagnostic investigation of unexpected serology results for swine influenza virus (SIV) and porcine reproductive and respiratory syndrome virus (PRRSV)
Kurt D. Rossow, DVM, PhD; Paul Yeske, DVM, MS; Sagar M. Goyal, DVM; Richard Webby, PhD; James E. Collins, DVM, PhD
KDR, SMG, JEC: Minnesota Veterinary Diagnostic Laboratory, St Paul, Minnesota; PY: Swine Vet Center, St Peter, Minnesota; RW: St Jude Children’s Hospital, Memphis, Tennessee.
Swine influenza virus (SIV) and porcine reproductive and respiratory syndrome virus (PRRSV) are two common pathogens recognized to exist within swine populations in multiple genomic forms.1,2 Serologic tests to diagnose pigs infected with SIV or PRRSV are commonly used by veterinary practitioners. Although interpretation of SIV and PRRSV serology results is generally straightforward, unexpected initial results sometimes occur, and may eventually provide insight for explaining therapeutic failures and considerations for future testing and result interpretation.Rossow KD, Yeske P, Goyal SM, et al. Diagnostic investigation of unexpected serology results for swine influenza virus (SIV) and porcine reproductive and respiratory syndrome virus (PRRSV). J Swine Health Prod. 2003;11(1):33-35.
SIV infection
In November 2001, tissues were submitted to the Minnesota Veterinary Diagnostic Laboratory (St Paul, Minnesota) to investigate an episode of coughing in a group of 15-week-old pigs in southern Minnesota. Eighty percent of 4000 pigs were affected. An influenza virus was isolated from tissues and was further characterized by standard hemagglutinin typing as an H3 influenza virus. The isolate was forwarded to St Jude Children’s Hospital (Memphis, Tennessee), where it was identified as a subtype H3N2 virus genetically similar to other central United States H3N2 viruses. Thirty convalescent sera obtained from this herd 4 weeks after the onset of clinical disease were initially tested by the hemagglutination inhibition (HI) test for antibodies against the standard H3N2 virus (A/SW/TX/98). Reciprocal titers >=40 were considered specific.3 Surprisingly, low (range 20 to 40) or no detectable titers were found (Table 1). In addition, all sera were negative for antibodies to the classic swine H1N1 virus. Subsequently, these convalescent sera were tested for antibodies against two other H3N2 influenza viruses: A/SW/WEB/87, a European H3N2 virus, and A/SW/MN/01, the H3N2 virus isolated from the affected pigs (homologous virus). Convalescent sera had expected titers (range 20 to 320) when tested by HI using the homologous H3N2 virus as antigen, while there was only a weak response (range 10 to 40) to the standard H3N2 virus (Table 1).
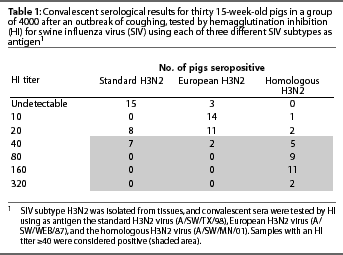
These HI results show that at least one H3N2 influenza virus in the US swine population does not react serologically with the prototype H3N2 influenza virus used routinely in diagnostic laboratories. These results also demonstrate the value of testing convalescent sera with homologous virus when serology results are unexpectedly negative. The HI titers to the homologous virus in this case are typical for convalescent samples 4 weeks post infection (personal observation). In this case, serologic testing using the homologous SIV verified the subtype of influenza virus isolated. The virus subtyping results may otherwise have been treated as suspect because of the apparent lack of seroconversion in the herd. Subsequent testing for H3N2 influenza virus in other groups of pigs from this herd may also require use of the homologous virus.
In some cases, influenza virus serology results may depend on the SIV strain used as antigen in the HI test. Diagnostic laboratories and practitioners should consider this when interpreting results. To our knowledge, the H3N2 virus in this case has not been identified in other swine herds.
PRRSV infection
A PRRSV-positive and vaccinated sow herd in southern Minnesota experienced a severe episode of abortion beginning in early May 2002. The sow herd was naive to this strain of PRRSV, on the basis of several years of PRRSV open reading frame (ORF) 5 sequence documentation. Sixty sera were submitted to the Minnesota Veterinary Diagnostic Laboratory for a diagnostic evaluation. Thirty-five sow sera and 15 nursery pig sera were tested for PRRSV antibodies by ELISA (PRRS HerdChek, Idexx Laboratories, Westbrook, Maine) and indirect fluorescent antibody (IFA).4 Historically, PRRSV strain VR-2332, the parent strain of the Ingelvac modified-live vaccine virus (Boehringer Ingelheim/Nobl Laboratories, Sioux Center, Iowa), has been used as antigen in the US PRRSV IFA test. The European prototype PRRSV, known as Lelystad strain, is used in the European PRRSV IFA test at the Minnesota Veterinary Diagnostic Laboratory.
In the affected herd, the US PRRSV strain was detected by polymerase chain reaction (PCR) in pooled sera from nursery pigs and sows. Selected pooled sera were negative for European PRRSV by PCR. Open reading frame 5 of the PRRSV isolate was sequenced and the strain was identified as a wild-type PRRSV known to be circulating in southern Minnesota (on the basis of cumulative sequence data). The disparity between results of Idexx ELISA and IFA using VR-2332 and the Lelystad strains as antigen (Table 2) prompted further serological testing. Selected sera were submitted to Eric Nelson (South Dakota Animal Disease Research and Diagnostic Laboratory, Brookings, South Dakota) to test for PRRSV ORF 7 antibodies using an experimental ELISA technique. An attempt was made to isolate PRRSV for use in homologous virus IFA testing; however, isolation was unsuccessful. In place of homologous PRRSV, a wild-type PRRSV adapted to MARC-145 cells was selected as an alternative to PRRSV VR-2332.5 Serology results are shown in Table 2. There was an 81.8% concordance between the IFA test using the wild-type US PRRSV and the Idexx ELISA test, compared to a 4.5% concordance when VR-3223 was used as antigen in the IFA test.
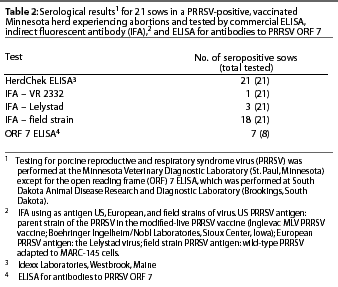
Infection with this US PRRSV demonstrated remarkable variation in the early antibody response as measured by IFA test. The disparity in an early antibody response by IFA test has been recognized for European PRRSV infections, but not for US PRRSV infections.6 As the immune response progresses, however, US and European antibodies measured by IFA testing begin to cross react with each other. Early antibodies in this infection did cross react with VR-2332 (one sample) and European PRRSV (three samples), but at a low concordance with the Idexx ELISA test (4.5% and 13.6%, respectively).
The results in this case have implications for routine PRRSV serologic testing. In potentially acute PRRSV infections, defining PRRSV ELISA results as false-positives on the basis of a negative IFA test should take into consideration the virus used in the IFA test. The field-type virus used in the IFA test in this case is probably more representative of the US PRRSV population than VR-2332, as the frequency of identification of VR-2332 wild-type viruses or vaccine viruses is low (Minnesota Veterinary Diagnostic Laboratory, unpublished data). However, there may also be other strains of PRRSV circulating in the US with similar properties. Using a battery of viruses as IFA antigens is not feasible on a daily basis in a diagnostic laboratory setting. Testing serum for PRRSV by PCR may provide additional important information for PRRSV serology interpretation in acute infections. In addition, the herd PRRSV history and monitoring program, clinical signs, and proximity to PRRSV-positive herds should all be considered when serology results are interpreted as false-positives or when a decision is made to conduct additional testing.
Conclusion
Unexpected serologic results should not be routinely dismissed as laboratory error or inappropriate sampling. They may provide opportunities to improve our understanding of the complexities of infectious disease and further assist animal health decisions.
References – refereed
1. Dee SA, Torremorell M, Rossow K, Mahlum C, Otake S, Faaberg K. Identification of genetically diverse sequences (ORF 5) of porcine reproductive and respiratory syndrome virus in a swine herd. Can J Vet Res. 2001;65:254-260.
2. Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon KJ, Krauss
S, Webster RG. Genetic reassortment of avian,
swine and human influenza A viruses in American pigs.
J Virol. 1999;73:8851-8856.
4. Yoon IJ, Joo HS, Christianson WT, Kim HS, Collins JE, Morrison RB, Dial GD. An indirect fluorescent antibody test for the detection of antibody to swine infertility and respiratory syndrome virus in swine serum. J Vet Diagn Invest. 1992; 4:144-147.
5. Bierk MD, Dee SA, Rossow KD, Collins JE, Guedes MI, Pijoan C, Molitor TW. Diagnostic investigation of chronic porcine reproductive and respiratory syndrome virus in a breeding herd of pigs. Vet Rec. 2001;148:687-690.
6. Bautista EM, Goyal SM, Collins JE. Serologic survey for Lelystad and VR-2332 strains of porcine respiratory and reproductive syndrome (PRRS) virus in US swine herds. J Vet Diagn Invest. 1993;5:612-614.
References – non refereed
3. Swenson SL. Swine influenza: serology and vaccination. Applied serology and vaccinology, Workshop 10. Ann Meet AASP. 1998;29:19-25