Original research |
Peer reviewed |
The impact of endemic porcine reproductive and respiratory syndrome virus and other pathogens on reproductive performance in swine
Gertraud Regula, DVM, MS; Gail Scherba, DVM, PhD; Nohra E. Mateus-Pinilla, DVM, PhD; Carol A. Lichtensteiger, DVM, PhD, Diplomate ACVP; Gay Y. Miller, DVM, PhD; Ronald M. Weigel, PhD
Department of Veterinary Pathobiology, College of Veterinary Medicine, University of Illinois, 2001 S Lincoln Avenue, Urbana, IL 61802. Corresponding author: Dr R. M. Weigel, Department of Veterinary Pathobiology, College of Veterinary Medicine, University of Illinois, 2001 S Lincoln Avenue, Urbana, IL 61802; Tel: 217-244-1365; Fax: 217-244-7421; E-mail: weigel@uiuc.edu.
Regula G, Scherba G, Mateus-Pinilla NE, et al. The impact of endemic porcine reproductive and respiratory syndrome virus and other pathogens on reproductive performance in swine. J Swine Health Prod. 2003;11(1):13-18. Also available as a PDF.
Summary
Objectives: To evaluate the impact of endemic infection with porcine reproductive and respiratory syndrome virus (PRRSV), swine influenza virus (SIV), transmissible gastroenteritis virus-porcine respiratory coronavirus (TGEV-PRCV), pseudorabies virus (PRV), Mycoplasma hyopneumoniae, and Actinobacillus pleuropneumoniae (APP) on the reproductive performance of sows.
Methods: Seventeen groups of 30 to 60 sows and gilts in seven herds were monitored over a 2-year period by serological testing for the pathogens listed above. Litter size, number of stillborn (including mummies), average weaning weight, preweaning deaths, and interfarrowing interval were recorded for the sows that were tested. Multiple linear regression analyses were performed to evaluate the association of the measures of reproductive performance with serological test results.
Results: Infection with PRRSV was consistently associated with poorer reproductive performance. Sows that had antibodies to PRRSV had, on average, 0.1 to 0.9 more stillborn piglets per litter than did seronegative animals. The average interfarrowing interval was 3 to 10 days longer for PRRSV-seropositive sows. A consistent correlation between reproductive performance and serological results could not be found for SIV, TGEV-PRCV, PRV, M hyopneu-moniae, or APP.
Implications: Even in herds where no clinical signs of the disease are present, reproductive performance may be substantially inferior in sows with high levels of antibody against PRRSV.
Keywords: swine, reproduction, endemic infection, porcine reproductive and respiratory syndrome virus
Received: July 19, 2001
Accepted: May 23, 2002
Pathogens such as porcine reproductive and respiratory syndrome virus (PRRSV), swine influenza virus (SIV), transmissible gastroenteritis virus (TGEV), Mycoplasma hyopneumoniae, and Actinobacillus pleuropneumoniae (APP) are prevalent in a large percentage of swine herds in the United States.1,2 Consequences of acute disease outbreaks due to these pathogens include abortions, stillbirths, reduced fertility of the sows, and increased mortality of the piglets.3-6 However, herds may be infected with these pathogens without showing signs that differentiate or characteristically define the presence of a specific disease.4,6,7 These endemic infections may be characterized by mild respiratory signs such as coughing or sneezing, or slightly reduced reproductive performance that does not elicit diagnosis and intervention by the producer.
Several studies have evaluated the financial impact of acute disease outbreaks in swine operations.5,8-10 The economic consequences of endemic infection, on the other hand, are more difficult to measure. For decisions regarding herd health management, it is crucial to have information on the impact of infection on productivity. Interventions such as vaccination or elimination of a pathogen from a herd must be evaluated against the consequences of allowing the herd to remain endemically infected. The objective of this study was to estimate the impact of endemic infections with PRRSV, SIV, TGEV, pseudorabies virus (PRV), M hyopneumoniae, and APP on reproductive performance. These pathogens were selected for monitoring because of their potential impact on productivity,4-6,11-14 and because a serological test that was economical and feasible for use under practice conditions was available for each organism. The impact of endemic infection with these pathogens on growth in finishing pigs has been evaluated previously.15
Materials and methods
Herds
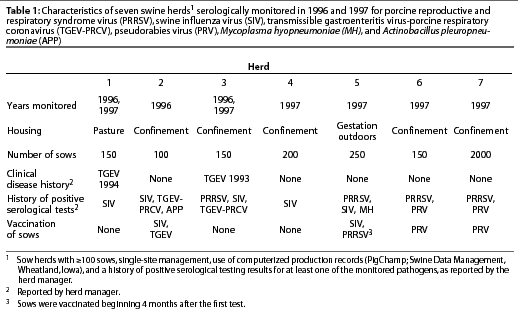
This study was conducted in herds that met the following criteria for inclusion: herd size >=100 sows, single-site management, use of computerized production records (PigChamp; Swine Data Management, Wheatland, Iowa), and a history of positive serological testing results for at least one of the monitored pathogens, as reported by the herd manager.
During the first year of the study, three university research herds were monitored, one of which (Herd 2) was depopulated at the end of the year. In that herd, only one group of 30 sows could be monitored before the farm was depopulated. In the second year, the two remaining and one new university research herd were monitored. Three commercial herds were also evaluated during 1997. A summary of herd characteristics and disease and vaccination history is provided in Table 1.
Sampling procedures
The study was conducted in 1996 and 1997, beginning in the spring of each year. Each herd was visited twice per year, with a 5- to 6-month interval between visits. In 1996, blood samples were collected from 30 sows in each herd, selected at random. This sample size provides a 95% probability of detecting seropositivity with a prevalence of at least 10%.16 The sample size was increased to 60 sows per herd in 1997, because many sows were lost to follow-up during the first year. With a maintained sample size of 60, there is a 95% probability of detecting seropositivity if the prevalence is at least 5%. Whenever possible, sows sampled during the first herd visit were re-sampled on the second visit, with the remainder required to achieve the desired sample size selected at random. Data on reproductive performance of sows sampled for serological testing were obtained from computerized production records. For each sampling, the reproductive performance of the tested sows was recorded for the farrowing preceding and following the serological test. The recorded measures of production were the number of piglets born alive, the number of piglets born dead (including mummies), the number of piglets fostered on or off, the number of pigs weaned, the weaning weight of the litter, and the time between two consecutive farrowings (interfarrowing interval).
Serological testing
Endemic infections with the following pathogens were monitored by serological assays performed at the University of Illinois Veterinary Diagnostic Laboratory (Champaign, Illinois): PRRSV, HerdChek PRRS ELISA (Idexx, Westbrook, Maine), positive cutoff sample-to-positive (S:P) ratio 0.4; SIV, hemagglutination inhibition test for serotype H1N1, positive cutoff titer 40; TGEV-porcine respiratory coronavirus (PRCV), serum neutralization (Purdue strain TGEV), positive cutoff titer 64; and M hyopneumoniae, ELISA, positive cutoff S:P ratio 0.6.
The HerdChek Anti-PRV g1 assay (Idexx), was performed at the Illinois Department of Agriculture Animal Disease Diagnostic Laboratory (Galesburg, Illinois), positive cutoff S:P ratio 0.7.
A complement fixation test for APP (serotypes 1, 3, 5, and 7) was performed at the Iowa State University Veterinary Diagnostic Laboratory (Ames, Iowa), positive cutoff titer 8.
Statistical methods
A difference in reproductive performance among seropositive and seronegative
animals of the magnitude of the standard deviation can be detected with a sample
size of 30 if the seroprevalence is 50% (power=0. 8 and
[alpha]=.05).16 With a lower or higher prevalence, only a
difference among groups that is somewhat larger than the standard deviation can be
detected with the same power, because, in this
case, the sample size in one group becomes smaller. To evaluate the impact of
endemic infection on sow productivity, multiple linear regression analyses were
conducted (PROC REG, SAS Institute, Cary, North Carolina). This statistical method
allows evaluation of the effect of a higher antibody titer to a pathogen on
reproductive performance, taking confounding
factors into account. The calculated effect is corrected for the influence of other
pathogens, as well as covariates such as parity of
the sow or group tested. The outcomes were interfarrowing interval, number of
liveborn piglets, number of stillborn piglets
(including mummies), preweaning deaths, and weaning weights for the litters
preceding and following the serological testing.
The average weaning weight per pig was calculated as the weight of the litter at
weaning divided by the number of pigs weaned. The number of preweaning deaths was
calculated as the number of piglets born alive plus (or minus) the number of piglets
fostered on (or off), minus the number of pigs weaned. All reproductive
measures except interfarrowing interval were recorded for the farrowing before and
the farrowing after the serological testing. The interfarrowing interval was calculated
from the farrowing before to the farrowing after the serological testing. Sows that did
not have records of two consecutive farrowings during the observation time were
excluded from the analysis of interfarrowing intervals. The data for each herd were
analyzed separately. In all of these models,
serological test results for each of the monitored infections were used as the predictors.
The proportion of variance accounted for uniquely by each predictor after all
other variables in the model had been taken into account, the squared semipartial
correlation coefficient (sr2), was calculated.
Serological test results were entered into the analyses as titer values or ELISA S:P
ratios rather than as positive and negative values in order to utilize more precise
quantitative information for the regression models.
Parity was included as a covariate in all regression models because it has been reported
to influence reproductive
performance.17 In Herd 5, vaccination status differed
between the two groups of sows tested. In this
herd, the product of vaccination status and serological titer was included as an
interaction variable. Regression models were tested
for satisfaction of the assumptions of normality, homogeneity of variance, the
presence of influential observations, and independence of
residuals.18 The method of variable selection was initial forced entry of
all predictors, with stepwise backward elimination. In order to control for
confounding in the model, the P-to-remove
criterion was set to .2. Thus, all variables with a
P-value <.2 were kept in the model. The level of significance was set to
[alpha]=.05. Predictions were that higher
serological titers would be associated with fewer
liveborn piglets, more stillborn piglets and mummies, more preweaning deaths,
a lower average weaning weight, and a longer interfarrowing interval. In vaccinated
animals, higher antibody levels might be associated with either a strong immune
response to vaccination or an infection with the pathogen. Therefore, no
predictions were made for vaccinated animals
unless the serological test was capable of distinguishing field virus from vaccine
virus.
Results
Reproductive performance of the study herds is summarized in Table 2.
Serological test results for the monitored herds are
presented in Table 3. Six of the seven herds were seropositive for PRRSV. In all
herds tested, seroprevalence of both SIV and
M hyopneumoniae was high. In five herds, more than 50% of sows tested positive
for TGEV-PRCV. There was a low prevalence of APP in three herds. In two herds,
there was a history of prior infectionwith PRV: no animals were sero-positive in one
of these herds, and only a few were seropositive in the other.
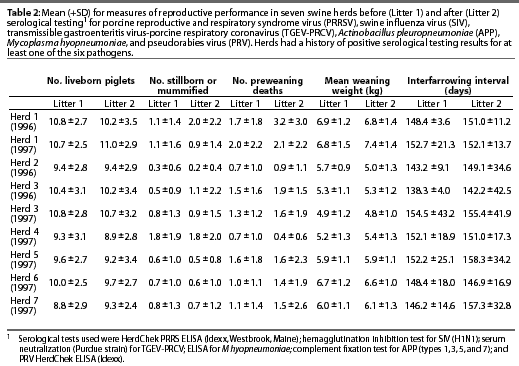
The most consistent associations between measures of reproductive performance and serology results detected were for PRRSV, which was associated with lower measures of reproductive performance in five of the seven herds. In Herd 6, higher PRRSV ELISA S:P ratios were correlated with fewer piglets born alive (sr2=0.06, P=.009). A sow with an S:P ratio of 0.4 (positive cutoff) had, on average, 0.34 fewer liveborn piglets per litter than a sow with an S:P ratio of 0. An S:P ratio of 1.41 (75th percentile) in Herd 6 was associated with an average of 0.72 fewer piglets compared to a ratio of 0.56 (25th percentile). In contrast, in Herd 5, in which one group of sows was vaccinated, a higher PRRSV ELISA S:P ratio was associated with more piglets born alive (sr2=0.09, P=.01). Furthermore, in Herds 3, 4, and 5, a higher PRRSV S:P ratio was associated with more stillborn and mummified piglets. In Herd 3 (1996), an S:P ratio of 0.4 (positive cutoff) was associated with an average of 0.52 more stillborn piglets and mummies compared to an S:P ratio of 0 (sr2=0.12, P=.004). There was a difference of 1.24 stillborn piglets and mummies per litter between sows with an S:P ratio=0.08 (25th percentile) and those with an S:P ratio=1.03 (75th percentile). This effect was smaller in 1997, when seropositive sows had, on average, only 0.14 more stillborn piglets and mummies than seronegative sows (sr2=0.04, P=.049). All sows in Herd 4 were seronegative for PRRSV. Nevertheless, sows with an S:P ratio of 0.04 (75th percentile) had an average of 0.90 more stillborn piglets and mummies per litter than sows with an S:P ratio of 0 (25th percentile) ( sr2=0.15, P=.004). In Herd 5, only the second group of sows was vaccinated against PRRSV. The effect of PRRSV S:P ratio on the number of stillborn pigs and mummies differed between the vaccinated and non-vaccinated groups. There was a marked increase in the number of stillborn piglets and mummies with a higher PRRSV S:P ratio in unvaccinated sows (sr2=0.10, P=.003). In vaccinated sows, the number of stillborn piglets increased only slightly with higher S:P ratios.
In Herd 1 (1996), piglets of sows that were seropositive for PRRSV were more likely to die between birth and weaning than were piglets from seronegative sows (sr2=0.02, P=.045). In Herd 5, a higher PRRSV S:P ratio was associated with a lower average weaning weight (sr2=0.06, P=.018). In Herd 2, in contradiction to the predicted direction of the effect, higher PRRSV S:P ratios were associated with higher average weaning weight (sr2=0.24, P=.016). In Herds 3, 4, and 5, a higher PRRSV S:P ratio was associated with a prolonged interfarrowing interval. In 1997, the interfarrowing interval for sows with a PRRSV S:P ratio of 0.4 (positive cutoff) was 4.6 days longer in Herd 5 (sr2=0.09, P=.014) and 10.4 days longer in Herd 3 (sr2=0.05, P=.036), compared to sows with an S:P ratio of 0. The difference in interfarrowing interval between sows with the 25th and 75th percentile PRRSV S:P ratios in 1997 was 10.9 days in Herd 3 and 24.7 days in Herd 5. In Herd 4, although all animals were negative for PRRSV, higher PRRSV S:P ratios were correlated with a longer interfarrowing interval (sr2=0.15, P=.005). Sows with an S:P ratio of 0.04 (75th percentile) on average had an interfarrowing interval 2.6 days longer than sows with an S:P ratio of 0 (25th percentile).
A consistent negative association with reproductive performance across herds could not be determined for any of the other monitored pathogens. Endemic SIV infection was associated in Herd 6 (1997) with fewer piglets born alive (sr2=0.12, P=.001). Sows seropositive for SIV in Herd 1 (1996) had, on average, fewer stillborn pigs (sr2=0.05, P=.034) than seronegative sows. In Herd 3 (1997), higher SIV titers were associated with more preweaning deaths (sr2=0.07, P=.011), while in Herd 1 (1996), higher SIV titers were associated with fewer preweaning deaths (sr2=0.16, P=.001).
Higher titers for TGEV-PRCV were associated with poorer production in some herds, and with better production in others. In Herd 6 (1997), litters from sows with higher TGEV-PRCV titers had fewer liveborn piglets than seronegative sows (sr2=0.03, P=.042). In Herd 1 (1997), higher TGEV-PRCV titers were correlated with higher preweaning mortality (sr2=0.11, P=.002). In Herd 2, the sows were vaccinated for TGEV, but there were more preweaning deaths in litters from sows with higher TGEV-PRCV titers (sr2=0.17, P=.009). In Herds 3 (1996) and 6 (1997), sows were not vaccinated against TGEV, and higher TGEV-PRCV titers were associated with fewer preweaning deaths (Herd 3: sr2=0.11, P=.048; Herd 6: sr2=0.07, P=.02). The average weaning weight was lower in litters from sows with higher TGEV-PRCV titers in Herd 1 (1997) (sr2=0.21, P<.001), as well as in Herd 2 (1996) (sr2=0.21, P=.022). The interfarrowing interval in litters from sows with higher TGEV-PRCV titers was longer in Herd 3 (1997) (sr2=0.10, P=.005) and shorter in Herd 6 (1997) (sr2=0.10, P=.018).
Actinobacillus pleuropneumoniae infection was associated with more preweaning deaths in Herd 2 (1996), which had the highest prevalence of this pathogen (sr2=0.29, P=.002).
The number of piglets born alive decreased with an increasing PRV S:P ratio (sr2=0.03, P=.045) in Herd 6 (1997), one of the two herds where PRV was present.
Mycoplasma hyopneumoniae, although prevalent in all herds, had an impact on reproductive performance only in Herd 6 (1997). Preweaning deaths increased (sr2=0.05, P=.022) and average weaning weight decreased (sr2=0.05, P=.027) with an increasing S:P ratio for M hyopneumo-niae in this herd.
Discussion
In this study, the impact of endemic infection on reproductive performance was measured in university herds and in a convenience sample of commercial herds. The data on pathogen prevalence are therefore not necessarily representative of Illinois swine herds.
Although there were no apparent clinical signs due to infection with PRRSV reported in any of these herds at the time of our study, serological evidence for infection with PRRSV was associated with poorer reproductive performance in five of the seven monitored herds. This virus is known to persist in the herd for a prolonged period of time.7 The effect of endemic infection with PRRSV on the monitored herds was of a magnitude that might have a substantial economic impact.
The magnitude of the effect of PRRSV infection on reproductive performance differed among herds. This may have been due to variations in pathogenicity of the virus and the immune status of the herds. Co-infection with other pathogens might also have influenced the effect of PRRSV on reproductive performance.
The negative association of endemic PRRSV infection with reproductive performance has been identified previously.14 The study of Baysinger et al14 agreed with this study in identifying fewer liveborn pigs, higher pre-weaning mortality, and a longer interfarrowing interval in herds classified by producers as subclinically infected, compared to ‘negative’ herds, and differed in finding no association of endemic PRRSV with stillborn births and mummies. Direct comparisons are difficult because in the present study, the individual sow was the unit of analysis, whereas Baysinger et al14 compared mean differences between herds, and did not require serological testing to confirm a herd as ‘negative’ for PRRSV infection.
There are several possible explanations for the presence of a significant correlation between higher PRRSV S:P ratios and more births of stillborn piglets and mummies and a longer interfarrowing interval in Herd 4, where no sows were diagnosed as serologically positive for PRRSV. First, it is possible that the positive cutoff value for the PRRSV serological test might be set too high to detect endemic infection. Second, the diagnostic test for PRRSV might have cross-reacted with another pathogen that had a negative effect on measures of reproductive performance. Finally, the significant association between PRRSV S:P ratios and reproductive performance might have occurred by chance.
In Herd 5, a different effect of PRRSV ELISA S:P ratio could be demonstrated for vaccinated and non-vaccinated sows. Even though the better reproductive performance in the vaccinated group might have been due to chance, this seems unlikely if one looks at the relationship between PRRSV S:P ratio and number of stillborn piglets and mummies per sow. In non-vaccinated sows, higher PRRSV S:P ratios were associated with a greater number of stillborn piglets and mummies. This relationship did not occur in vaccinated sows.
With the exception of PRRSV, associations of a specific endemic infection and poorer reproductive performance could be found only in some herds. Some effects occurred in the opposite direction to that predicted, especially for preweaning deaths and weaning weight. There were repeated significant associations of TGEV-PRCV with reproductive performance, but some results were contradictory. Several different factors might be responsible for the conflicting results. First, there was a large variation in reproductive performance among individual sows, due in part to differences in genetic potential and in part to random variation.17 Second, an endemic infection with a pathogen might have a negative impact only under certain management conditions, and the effect of a single pathogen might also be dependent on general herd health status and the strain of the organism.19 Third, the time when each herd was infected with the monitored pathogens was unknown. A negative association of antibody levels with reproductive performance may be expected if the pathogen persists in the animal, if the infection occurs during the observation period, or if previous infection has caused morphological damage to the reproductive system, thereby having a negative impact on reproductive performance beyond the period of infection and persistence of antibodies. Immunity of the sows from a prior infection might have had a positive impact on reproductive performance: high antibody levels of the sow might have reduced preweaning deaths and increased average weaning weight through the protective effect of maternal antibodies.
In any study of the association of multiple diagnostic test results with measures of reproductive performance, some associations might be significant by chance. The proportion of variance accounted for by some predictors was small and may not be biologically important. With the sample size of sows used in most analyses (n = approximately 40), there was a statistical power of 0.80 to detect an sr2 value of at least 0.14 as significant at [alpha]=.05 in a one-tailed test.16 Therefore, the association of any single variable with performance should not be overinterpreted. Repeated strong associations over time in a herd identifies a problem specific to that herd. It is valid to generalize the results to other herds than the ones that participated in our study only if an association has been identified in multiple herds. In this study, this was the case only for PRSSV, ie, only PRRSV infection showed a consistent, strong association with inferior reproductive performance across herds.
Implications
- Endemic PRRSV may have a signifi-cant negative impact on reproductive performance of sows.
- Measuring differences in performance between seropositive and seronegative sows within one herd may give an estimate of the effect of infection in that herd, and may also show the benefit of vaccinating sows.
- Decisions on herd health management may thus be based on economical analysis of costs and benefit of intervention.
Acknowledgment
Supported by a grant from the Illinois Council on Food and Agricultural Research.
References – refereed
3. Pejsak Z, Stadejek T, Markowska-Daniel I. Clinical signs and economic losses caused by porcine reproductive and respiratory syndrome virus in a large breeding herd. Vet Microbiol. 1997;55:317-322.
4. Easterday BC. Swine influenza. In: Leman AD, Straw BE, Glock RD, Mengeling WL, Penny RHC, Scholl E, eds. Diseases of Swine. 6th ed. Ames, Iowa: The Iowa State University Press; 1986:244-255.
5. Miller GY, Kliebenstein JB. The economic impact of clinical transmissible gastroenteritis for swine producers participating in the Missouri mail-in-record program. Prev Vet Med. 1983;3:475-488.
6. Ross RF. Mycoplasmal diseases. In: Leman AD, Straw BE, Glock RD, Mengeling WL, Penny RHC, Scholl E, eds. Diseases of Swine. 6th ed. Ames, Iowa: The Iowa State University Press; 1986:469-483.
7. Zimmerman JJ, Yoon KJ, Wills RW, Swenson SL. General overview of PRRSV: A perspective from the United States. Vet Micro. 1997;55:187-196.
8. Mullan BP, Davies GT, Cutler RS. Simulation of the economic impact of transmissible gastroenteritis on commercial pig production in Australia. Aust Vet J. 1994;71:151-154.
9. Poulson D, Marsh WE, Morrison RB, Dial G. A methodology for evaluating the financial consequences of a disease outbreak of transmissible gastroenteritis and pseudorabies virus. Prev Vet Med. 1993;16:61-63.
10. Rougoor CW, Dijkhuizen AA, Huirne RBM, Marsh WE. Impact of different approaches to calculate the economics of disease in pig herding. Prev Vet Med. 1996;26:315-328.
11. Christianson WT, Joo HS. Porcine reproductive and respiratory syndrome: A review. Swine Health Prod. 1994;2:10-28.
12. Parsons TD, Pitcher PM, Johnstone C. Economic analysis of an epizootic of pseudorabies and subsequent production following the institution of a vaccine program in a Pennsylvania swine herd. JAVMA. 1990;197:188-191.
13. Fedorka-Cray PJ, Anderson GA, Cray WC, Grey JT, Breisch SA. Actinobacillus (Haemophilus) pleuropneumoniae. Part II. Virulence factors, immunity, and vaccines. Comp Cont Ed Pract Vet. 1994;16:117-120.
14. Baysinger AK, Dewey CE, Straw BE, Brumm MC, Schmitz J, Doster A, Kelling C. The effect of PRRSV on reproductive parameters in swine herds. Swine Health Prod. 1997;5:173-176.
15. Regula G, Lichtensteiger CA, Mateus-Pinilla NE, Scherba G, Miller GY, Weigel RM. Comparison of serologic testing and slaughter evaluation for assessing the effects of subclinical infection on growth in pigs. JAVMA. 2000;217:888-895.
16. Beal VC. Regulatory Statistics. Vol 1. 6th ed. United States Department of Agriculture, Animal and Plant Health Inspection Service, Veterinary Services; 1983:19-21.
17. Whittemore C. The Science and Practice of Pig Production. Essex, United Kingdom: Longman Scientific and Technical; 1993.
18. Neter J, Wasserman W, Kutner MH. Applied Linear Statistical Models: Regression, Analysis of Variance, and Experimental Designs. Homewood, Illinois: Irwin; 1985.
19. Goldberg TL, Weigel RM, Hahn EC, Scherba G. Associations between genetics, herd characteristics and clinical disease in field outbreaks of porcine reproductive and respiratory syndrome virus. Prev Vet Med. 2000;43:293-302.
References – non refereed
1. National Swine Survey: Morbidity/mortality and health management of swine in the United States. Fort Collins, Colo: Centers for Epidemiology and Animal Health, Animal and Plant Health Inspection Service; 1992. United States Department of Agriculture Publication N101. 0192.
2. Swine ’95: Grower/Finisher, Part I: Reference of 1995 swine management practices. Fort Collins, Colo: Centers for Epidemiology and Animal Health, Animal and Plant Health Inspection Service; 1995. United States Department of Agriculture Publication N186. 995