Oliveira S. 2002;5:221-225 Diagnosis of Haemophilus parasuis in
Diagnostic notes |
Non refereed |
Diagnosis of Haemophilus parasuis in affected herds and use of epidemiological data to control disease
Simone Oliveira, DVM, MS; Carlos Pijoan, DVM, MS, PhD
Department of Clinical and Population Sciences, University of Minnesota, St Paul MN 55108. E-mail: oliv0107@tc.umn.edu.
Oliveira S, Pijoan C. Diagnosis of Haemophilus parasuis in affected herds and use of epidemiological data to control disease. J Swine Health Prod. 2002;10(5):221-225.
Haemophilus parasuis is a commensal organism in the upper respiratory tract of pigs.1 This organism is particularly interesting due to its ability to invade the host and cause severe lesions characterized by fibrinous polyserositis, arthritis, and meningitis. Little is known about the pathogenesis, virulence factors, and immunogenicity of H parasuis, which makes control of systemic infections a difficult task.
Diagnosis of Haemophilus parasuis as a primaryagent causing disease in a herd requires evaluation of the herd’s health status and association of H parasuis with characteristic clinical signs and lesions. Control of disease, on the other hand, depends on characterization of the strains involved in systemic disease and an understanding of the epidemiology of the agent within and between herds. In this article, we will discuss how to improve H parasuis diagnosis in affected herds, how systemic strains may be characterized, and how strain-characterization data may be used in disease control.
Clinical characterization
and lesions
Haemophilus parasuis usually affects young pigs after 4 to 6 weeks in the nursery. Affected animals may show a variety of clinical signs and lesions, depending on the immune status of the herd, virulence of the strain, and stage of infection. Systemic infection by H parasuis in naive animals is characterized clinically by high fever (41.7°C), lethargy, swollen joints, coughing, abdominal breathing, and central nervoussystem signs, such as trembling and paddling. Lesions are characterized by fibrinous exudate on the pericardium, pleura, peritoneum, meninges, and joints.2 These clinical signs and lesions generally occur when highly virulent H parasuis strains are introduced into a susceptible herd, or when concurrent infections, for example, porcine reproductive and respiratory syndrome virus (PRRSV), are affecting the herd.
In endemically infected, stable herds,
H parasuis usually affects older animals. In these herds,
occurrence of disease is more sporadic and less severe than in
naive populations, and lesions may be restricted to the lung.
Pneumonia is characterized by antero-ventral consolidation, with
purulent exudate in the bronchi and bronchioli. Occasionally,
arthritis may occur in adult populations, especially in sow herds.
Genotyping and serotyping
H parasuis isolates
Haemophilus parasuis field isolates may be characterized by serotyping or genotyping. Serotyping is based on the reaction between serovar-specific polyclonal antiserum and heat-stable antigens extracted from the bacterial cultures, either in an agar gel precipitation test (AGPT) or an enzyme-linked immunosorbent assay (ELISA).3,4 Genotyping of H parasuis isolates has been performed by repetitive-element-based PCR (Rep-PCR).5,6,7
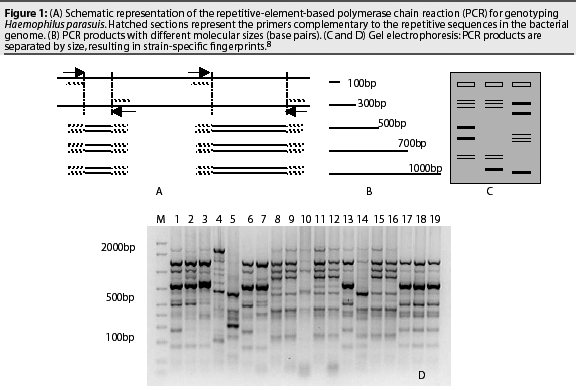
The Rep-PCR is a molecular-based technique that uses repetitive sequences within the bacterial genome (enterobacterial repetitive intergenic consensus (ERIC) sequences) to produce strain-specific fingerprints. The ERIC-PCR technique is schematized in Figure 1. Primers targeting the ERIC sequence align to the bacterial DNA and new double-stranded DNA is produced (Figure 1A). The repetitive sequences are randomly distributed in the bacterial genome, and the distance between two repetitive sequences may vary between different strains. This results in the production of several amplicons with different sizes (Figure 1B). The PCR products are then separated by size through electrophoresis in an agarose gel (Figure 1C). The resulting DNA pattern is called a fingerprint, and the process is called genotyping. Similar strains show comparable DNA bands, while different strains have varied distribution of the DNA bands (Figure 1D).8
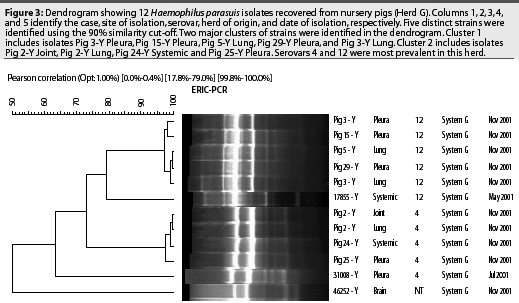
Fifteen serovars of H parasuis have been identified using the AGPT.9 A recent epidemiological study characterized 98 H parasuis field isolates recovered from 15 North American herds between 1999 and 2001.10 Isolates were serotyped using the AGPT and genotyped using the ERIC-PCR technique. Serovar 4 (38.8%) and non-typable serovars (27.6%) were the most prevalent among the evaluated isolates. Other serovars that were identified included serovars 3 (8.2%), 1 (7.1%), 12 (7.1%), 2 (4.1%), 14 (3.1%), 5 (2%), 7 (2%) and 13 (1%). The ERIC-PCR technique detected a high genetic diversity within serovars. Twelve different strains were identified among isolates from serovar 4 (Figure 2). Non-typable isolates were divided into 18 genotypes. Genetic diversity was also observed within isolates from serovars 1, 3, and 7 (two strains each), serovars 12 and 14 (three strains each), and serovar 2 (four strains).
These results demonstrate the limitations of serotyping for characterizing and differentiating H parasuis strains. However, serotyping may still be useful for selection of commercial vaccines. Haemophilus parasuis isolates of the same serovar may induce satisfactory homologous protection, while heterologous protection is restricted or absent.11,12 Commercial vaccines generally produce inconsistent results between herds, possibly because prevalent serovars differ among herds. Another hypothesis is that different strains of the same serovar do not induce cross-protection. Although H parasuis is highly prevalent among US herds, different herds are infected by different strains (Figure 2), and it is unknown whether or not a commercial vaccine containing a unique strain can protect herds affected by heterologous strains of the same serovar. Further studies are necessary in order to test this hypothesis.

Genotyping provides an accurate characterization of any H parasuis isolate, including non-typable isolates, and differentiates between strains of the same serovar. This information is very useful for selection of strains to be included in autogenous vaccines. Moreover, genotyping may be used to compare H parasuis strains isolated from recipient and supplier herds and to monitor the introduction of new strains into the herd.8
Diagnosis of H parasuis: Selection of animals and tissues for sampling
Selection of appropriate animals and tissues for sampling is essential for the correct diagnosis of H parasuis. Sample animals that are showing characteristic signs of H parasuis infection. Better results are obtained if samples are collected from animals with both respiratory signs and arthritis. Select animals in the early stages of infection (newer cases) that have not been treated with antibiotics for at least 1 week. Euthanize clinically affected animals and using a swab, sample the fibrinous exudate covering the surface of the pericardium, pleura, and peritoneum, or joint and cerebrospinal fluids. Sampling dead animals significantly reduces the chances of isolating H parasuis. Keep samples refrigerated at 4°C until shipment and submit them to a diagnostic laboratory as soon as possible, using ice packs and a Styrofoam container.
Haemophilus parasuis can be isolated from the nasal cavity, tonsils, and trachea of healthy animals. Occasionally, H parasuis may be isolated either from healthy lungs or lungs with severe pneumonia lesions. Major differences exist among strains of H parasuis isolated from the upper respiratory tract, from lung samples, and from systemic sites. Strains that cause systemic infection have a low prevalence in the upper and lower respiratory tracts. Isolates from nasal cavities and tonsillar areas are particularly diverse serotypically and genotypically, and are usually of low virulence. Lung isolates are less diverse than nasal isolates, but may show a higher genetic variability than systemic strains. Strains isolated from pneumonic sites may or may not include the strains that are actually causing disease in the herd.7 Considering these factors, only samples from non-respiratory tract sites should be submitted for H parasuis isolation and diagnosis.
Interpreting results and deciding on disease control
Once H parasuis is isolated from affected animals, evaluate herd health status to decide whether H parasuis is a primary or a secondary problem. Herds experiencing an active PRRSV infection may have a significant increase in nursery mortality due to secondary agents such as Streptococcus suis and H parasuis. Until PRRSV infection is controlled, management of secondary agents through vaccination is generally unsuccessful. In some herds, stabilizing PRRSV infection is the key to H parasuis control.
When H parasuis is isolated from systemic sites in diseased nursery pigs in a PRRS-stable herd, efforts should be concentrated on disease prevention through vaccination or treatment. Control of H parasuis depends on understanding the epidemiology of the agent within and between herds.8 The genetic variability of systemic isolates is generally limited within herds, with only one or two strains involved in systemic disease.7 Characterization of strains isolated from related herds (supplier and recipient) is very useful for identifying potential sources of pathogenic strains. In some herds, use of either commercial or autogenous vaccines controls disease. Commercial vaccines contain several strains of different serovars, including non-typable strains. Autogenous vaccines usually include one or two prevalent strains recovered from the affected herd. In herds experiencing mortality due to H parasuis, try to characterize as many isolates as possible by serotyping and genotyping, and continue to do so as new cases occur after the vaccination program has been implemented. Serotyping is useful for selection of commercial vaccines, while genotyping can be used to select prevalent strains to be included in autogenous vaccines. After prevalent pathogenic strains have been characterized, decide whether to use a commer- cial or an autogenous vaccine. Either provides satisfactory results if the vaccine strains are similar to those causing disease in the affected herd, but heterologous protection is very limited. Non-typable isolates may be responsible for severe outbreaks, and it is difficult to match them with vaccine strains. It is not known how the high genetic diversity within serovars and within non-typable isolates relates to cross-protection. If commercial vaccines are not effective, use of autogenous vaccines should be considered. Strains to be included in autogenous vaccines should be the prevalent genotypes affecting the herd.
Haemophilus parasuis isolates genotyped by Rep-PCR can be compared using dendrograms (Figure 3), which may be constantly updated with new isolates as new cases appear in the herd. Clusters of prevalent strains are easily identified. Autogenous vaccines should include a representative strain from each major cluster. Always select strains isolated from systemic sites, such as pericardium, pleura, peritoneum, joints, or meninges.
Timing of vaccination is very important. Implementation of an appropriate vaccination protocol will depend on the epidemiology of the disease in the herd. Although disease caused by H parasuis may be observed as early as 2 weeks after placement in the nursery, most affected herds will have a peak of infection after 4 to 6 weeks in the nursery, when maternal immunity has declined and previously exposed animals infect naive animals. In herds experiencing disease late in the nursery, vaccinate at weaning and again 2 weeks later. In herds experiencing significant death losses soon after weaning, vaccinate prior to weaning. Although sows may also be vaccinated, results are better with pig vaccination. When sows are vaccinated, maternal immunity in pigs may persist until 6 to 7 weeks of age, and interferes with development of active immunity. Never vaccinate sows and piglets at the same time.
Summary
Control of H parasuis depends on correct diagnosis of the agent, accurate characterization of the strains causing systemic disease in the herd, and selection of an appropriate vaccine and vaccination protocol. Understanding the epidemiology of H parasuis in a particular swine population is still the key to control in affected herds.
References – refereed
1. Møller K, Kilian M. V factor-dependent members of the family Pasteurellaceae in the porcine upper respiratory tract. J Clin Microbiol. 1990;28:2711-2716.
2. Vahle JL, Haynes JS, Andrews JJ. Experimental reproduction of Haemophilus parasuis infection in swine: clinical, bacteriologic, and morphologic findings. J Vet Diagn Invest. 1997;7:476-480.
3. Morozumi T, Nicolet J. Some antigenic properties of Haemophilus parasuis and a proposal for serological classification. J Clin Microbiol. 1986;23:1022-1023.
5. Versalovic J, Schneider M, de Bruijn FJ, Lupski JR. Genomic fingerprinting of bacteria using repetitive sequence based PCR (rep-PCR). Meth Cell Mol Biol. 1994;5:25-40.
6. Rafiee M, Bara M, Stephens CP, Blackall PJ. Application of ERIC-PCR for the comparison of isolates of Haemophilus parasuis. Aust Vet J. 2000;78:846-849.
7. Ruiz A, Oliveira S, Torremorell M, Pijoan C. Outer membrane proteins and DNA profiles in strains of Haemophilus parasuis recovered from systemic and respiratory sites. J Clin Microbiol. 2001;39:1757-1762.
9. Kielstein P, Rapp-Gabrielson VJ. Designation of 15 serovars of Haemophilus parasuis based on immunodiffusion using heat-stable antigen extracts. J Clin Microbiol. 1992;30:862-865.
12. Rapp-Gabrielson VJ, Kocus GJ, Clark JT, Muir SK. Haemophilus parasuis: immunity in swine following vaccination. Vet Med. 1997;92:83-90.
References – non refereed
4. Lin BC, Cobb S. A "fuzzy" ELISA for serotyping Haemophilus parasuis. Proc IPVS. Bankok, Thailand. 1994;156.
8. Oliveira S, Pijoan C. Haemophilus parasuis: Improvement of diagnosis by a molecular-based technique. Proc AASV. Nashville, Tennessee. 2001;479.
10. Oliveira S, Blackall P, Pijoan C. Characterization of Haemophilus parasuis isolates by serotyping and genotyping. Proc IPVS. Ames, Iowa. 2002;224.
11. Oliveira S, Pijoan C. Use of an uncapsulated Haemophilus parasuis vaccine in pigs. Proc AASV. Kansas City, Missouri. 2002;319.