Otake S. 2002;2:59-65 Transmission of PRRSV by fomites
Original research
Peer reviewed
Transmission of porcine reproductive and respiratory syndrome virus by fomites (boots and coveralls)
Satoshi Otake, DVM; Scott A. Dee, DVM, PhD, Diplomate ACVM; Kurt D. Rossow, DVM, PhD; John Deen, DVM, PhD, Diplomate ABVP; Han Soo Joo, DVM, PhD; Thomas W. Molitor, PhD; Carlos Pijoan, DVM, PhD
SO, SAD, JD, HSJ, TWM, CP: Center for Disease Eradication, University of Minnesota College of Veterinary Medicine, 385 Animal Science-Veterinary Medicine Building, 1988 Fitch Avenue, St Paul, MN 55108. KDR: University of Minnesota College of Veterinary Medicine, 1332 Gortner Avenue, St Paul, MN 55108.
Otake S, Dee SA, Rossow KD, et al. Transmission of porcine reproductive and respiratory syndrome virus by fomites (boots and coveralls). J Swine Health Prod. 2002;10(2):59-65. Also available as a PDF
The following errata were present in the original but have been corrected in this version (April 15 2002):
· The credentials of one of the authors, Dr John Deen, were erroneously reported as "DVM, PhD, Diplomate ACVM". Dr Deen’s correct credentials are "DVM, PhD, Diplomate ABVP".
· In Figure 1, page 61, the letter designates on the diagrams for the Alternative protocol and the Standard protocol were inadvertently reversed on the right side of the figure. The designate for the Standard protocol (Group 4) should be the letter "C", and for the Alternative protocol (Group 5), the designate should be the letter "D".
Summary
Objectives: To determine if porcine reproductive and respiratory syndrome virus (PRRSV) can be transmitted to susceptible pigs by fomites or people exposed to infected pigs.
Methods: Ninety-six 4-week-old pigs from a PRRSV-naive
source were organized into six groups individually housed in isolation
rooms (four replicate trials, 24 pigs per trial). Group 1 pigs
were inoculated intranasally with PRRSV strain VR-2332
(2 mL, 105 median tissue culture infective doses per
mL.) On days 5, 6, and 7 post inoculation, investigators exposed
to Group 1 pigs attempted to transmit PRRSV to sentinel pigs (Groups
2 to 5) by contact. After exposure to the infected pigs, an investigator
entered the Group 2 room (Direct Contact group) wearing contaminated
boots and coveralls and without washing hands. In contrast, investigators
who entered the rooms housing Groups 3 to 5 were required to complete
specific sanitation protocols, which included changing boots and
coveralls and washing hands (Danish System, Group 3); changing
boots and coveralls, showering, and 12 hours down time (Standard
Protocol, Group 4); and changing boots and coveralls and showering,
with no down time (Alternative Protocol, Group 5).
Results: The PRRSV was detected on contaminated coveralls, boots, and hands of investigators who had contacted Group 1 pigs. Transmission of PRRSV occurred betweenGroups 1 and 2, but not between Group 1 and Group 3, 4, or 5.
Implications: The PRRSV can be transmitted to susceptible pigs by contaminated fomites (boots and coveralls) and hands; however, the use of sanitation protocols appears to limit its spread.
Keywords:  swine, porcine reproductive
and respiratory syndrome virus, fomites, biosecurity
swine, porcine reproductive
and respiratory syndrome virus, fomites, biosecurity
Received: June 18, 2001
Accepted: August 20, 2001
Inanimate objects (fomites), such as contaminated boots, coveralls, and transport vehicles, have long been consideredto be risk factors for pathogen transmission in swine. Swine pathogens that have been recovered from fomites includepseudorabies virus (steel and rubber)1 and Streptococcus suis (truck tires).1,2 The capacity for people to transmit swine pathogens is not clear at this time, although foot-and-mouth disease virus, swine influenza virus, and Pasteurella multocida have been recovered from humansexposed to infected swine.3-5
Porcine reproductive and respiratory syndrome virus (PRRSV) is an economically significant pathogen of swine.6 Introduction of PRRSV into naive herds has been documented to occur through infected pigs and semen.7,8 The role of fomite transmission of PRRSV is not well understood; however, virus has been recovered from a number of experimentally contaminated surfaces, including stainless steel, plastic, and rubber.9 In a recently published report, no mechanical transmission of PRRSV was demonstrated when personnel exposed to experimentally infected pigs contacted susceptible pigs.10 In contrast, contaminated needles have been shown to transmit PRRSV from infected to susceptible pigs.11
Because all routes of PRRSV entry into naive herds are not known at this time, farm owners frequently require employees and visitors to comply with strict sanitation protocols prior to entry. These protocols, commonly referred to as "biosecurity protocols,"12 include changing clothing and footwear, showering into and out of the facility, and refraining from contact with swine for 12 to 72 hours before entering the premise. Despite their widespread acceptancein the industry, the scientific foundation for the need of such protocols is lacking.
In order to determine whether biosecurity protocols are important in preventing the spread of PRRSV, it is first essential to identify potential means of PRRSV transmission. Therefore, the objectives of this study were to evaluate the ability of contaminated fomites (boots and coveralls) to transmit PRRSV to naive pigs, and to determineif PRRSV could be detected on various anatomical sites of the investigators after they had contacted infected pigs, and before and after use of specific sanitation protocols commonly practiced in the swine industry.
Materials and methods
Animals, housing, and infection model
A total of 96 4-week-old pigs were purchased from a farm known to be PRRSV-negative on the basis of 5 years of diagnostic data and absence of clinical signs. Four replicate trials were conducted, with 24 pigs organized into six groups in each trial. The experimental design is summarized in Figure 1.
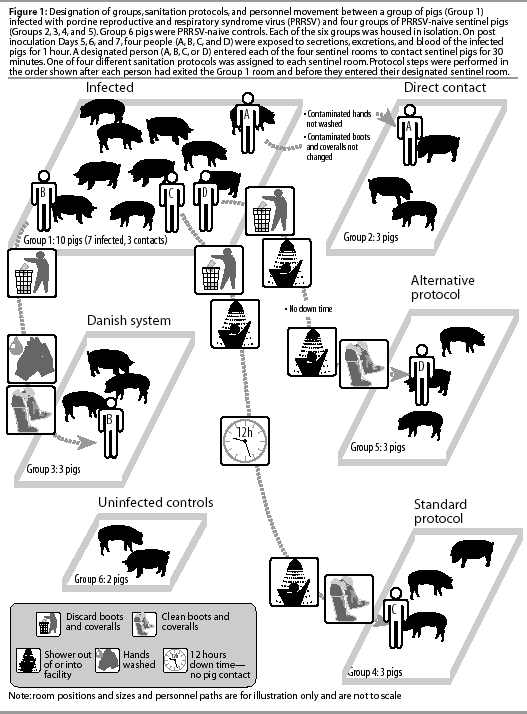
Each group was housed in a separate room in the isolation facility at the University of Minnesota College of Veterinary Medicine. The rooms were ventilated separately and contained individual slurry pits, minimizing the risk of cross-contamination between rooms. Each isolation room was providedwith an anteroom for hand washing and changing of boots and coveralls.
Group 1 (infected group) consisted of ten pigs; Groups 2, 3, 4, and 5 (sentinel groups) consisted of three pigs each; and Group 6 (negative control group) consisted of two pigs. Upon arrival at the isolation facility, all pigs were blood-tested by ELISA (IDEXX Laboratories, Westbrook, Maine)13 to confirm PRRSV-naive status. A sample-to-positive (S:P) ratio >=0.4 was considered positive. During the study period,pigs were cared for under the guidelines of University of Minnesota Institutional Animal Care policies.
To initiate each trial, seven of the ten pigs in Group 1 were inoculated intranasally with 2 mL of PRRSV (strain VR-2332). The remaining three pigs in Group 1 served as contact controls (Figure 1). The inoculum, prepared by passaging the virus five times on MARC-145 cells, was suspended in minimal essential medium (MEM) and administered at a concentration of 105 median tissue culture infective doses (TCID50) per mL.7
Group designations and protocols
Group designation is summarized in Figure 1. After experimental infection, attempts were made to transmit PRRSV from Group 1 (infected group) to Groups 2, 3, 4, and 5 (sentinel groups). A designated person (A, B, C, or D) was assigned to each sentinel group and was required to follow specific procedures after leaving the infected group and prior to entering the sentinel room.
Group 2 was designated the Direct Contact group, and the person entering the Group 2 room was designated Investigator A. After exposure to infected pigs in Group 1, Investigator A was required to wear contaminated boots (disposable plastic boots; First Priority Inc, Elgin, Illinois) and coveralls (Tyvek coveralls; Sunrise Industries, Albertville, Alberta), and to avoid washing hands after leaving the infected room and before entering the Group 2 sentinel room.
Group 3 was designated the Danish System group,12 and the investigator entering the Group 3 room was designated Investigator B. After exposure to infected pigs in Group 1, Investigator B was required to remove contaminated boots and coveralls, wash hands, and put on clean boots and coveralls prior to entering the Group 3 sentinel room.
Group 4 was designated the Standard Protocol group, and the investigator entering the Group 4 room was designated Investigator C. After exposure to infected pigs in Group 1, Investigator C was required to remove contaminated boots and coveralls, take a shower, and avoid contact with swine for a 12-hour period (down time). Investigator C was then required to shower into the isolation facility and to don clean boots and coveralls before entering the Group 4 sentinel room.
Group 5 was designated the Alternative Protocol Group, and the investigator entering the Group 5 room was designated Investigator D. The sanitation protocol for Investigator D was identical to the Standard Protocol, except that Investigator D showered out of the facility and immediately showered back in to enter the Group 5 room, with no down time.
Throughout the study, Investigators A, B, C, and D were in charge of feeding, cleaning, and sampling the animals in their designatedsentinel groups, and never entered the other sentinel rooms. Different personnel employed by the isolation facility were responsible for the care of Group 1 pigs throughout the study, and never entered the sentinel rooms. Group 6 pigs were cared for by isolation facility personnel, but were inspected at the beginning of each study day, prior to inspection of infected pigs.
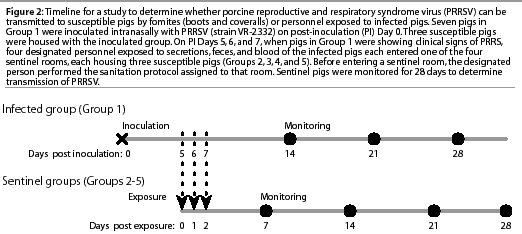
Study timeline
The study timeline is summarized in Figure 2. Attempts to infect sentinel Groups 2, 3, 4, and 5 took place on post-inoculation (PI) Days 5, 6, and 7. This exposure period was selected on the basis of published data documenting the occurrence of PRRSV transmission by contact between experimentally infected nursery pigs and control pigs.7
Protocol for exposure of investigators to infected pigs
During exposure to infected pigs in Group 1, investigators wore boots and coveralls, but did not wear hair nets, masks, or gloves. On each of the 3 days of the exposure period, investigators spent exactly 60 minutes in the Group 1 room to enhance contact with nasal secretions, saliva, blood, and fecal material from the infected pigs. Investigators picked up each pig and followed a standard operating protocol that was consistent across all trials. To contact nasal secretions, the surface of the snout was placed in contact with the dorsal and ventral surfaces of both hands and was rubbed repeatedly across the front surface of the boots and coveralls. To contact saliva,each hand was placed in the mouth of each pig for 5 seconds. To contact blood, 5 mL of whole blood collected from each pig in an EDTA tube (Vacutainer; Becton Dickinson, Franklin Lake, New Jersey) was drawn into a sterile 12-mL syringe(Monoject, St Louis, Missouri) and sprayed over the front surface of the coveralls, all outer surfaces of each boot, and the dorsal and ventral surfaces of the hands. To contact feces, a handful of fecal material was spread over both surfaces of the hands and on the boots and coveralls as described. In addition, investigators sat on the floor and allowed pigs to gently bite and lick their hands, coveralls, and boots.
Sanitation protocols
After the 60-minute contact period, investigators moved to designated sentinel rooms (Groups 2, 3, 4, and 5), performing the sanitation protocols summarized in Figure 1.
The Direct Contact person (Investigator A) removed contaminated boots and coveralls in the Group 1 anteroom and transferred them in a plastic bag to the Group 2 anteroom. Investigator A did not shower or wash hands after exiting the Group 1 room or before entering the Group 2 sentinel room, and dressed in the contaminated boots and coveralls to enter the sentinel room.
The Danish System person (Investigator B) removed contaminated boots and coveralls in the Group 1 anteroom, and moved to the Group 3 anteroom to wash hands and put on clean boots and coveralls before entering the sentinel room. The procedure of hand washing was as follows: contact with hot water (10 seconds), contact with soap (30 seconds), and rinsing with hot water (10 seconds).
After exposure to the infected Group 1 pigs, the Standard Protocol person (Investigator C) changed boots and coveralls in the Group 1 anteroom, showered out of the facility, and took a 12-hour period of down time (no contact with swine). After the down time, Investigator C showered into the facility, donned clean boots and coveralls, and entered the Group 4 sentinel room. The protocol of showering in and out included washing the hands (described above), washing the hair (shampoo, 60 seconds;rinsing, 60 seconds), and washing the body and face (soap, 100 seconds; rinsing, 100 seconds).
After exposure to the infected Group 1 pigs, the Alternative Protocol person (Investigator D) changed boots and coveralls in the Group 1 anteroom, showered out of the isolation facility, immediately showered back into the facility (no down time), and donned clean boots and coveralls to enter the Group 5 sentinel room.
Protocol for contact with sentinel pigs
Exactly 30 minutes of direct contact betweenthe designated investigator and the pigs in each sentinel group was required to enhance exposure to PRRSV contamination. The protocol of contact was similar to that described for the Group 1 pigs, except that there was no exposure to blood and feces of sentinel pigs. After the exposure period, investigators B, C, and D removed and disposed of their boots and coveralls in the sentinel anterooms, but the contaminated boots and coveralls of Investigator A were left in the Group 2 pen for the 24 hours after each exposure period. All investigators showered out of the facility after contact with the sentinel pigs.
Animal sampling
Sera were collected from all ten pigs in Group 1 on PI Days 2, 5, 6, 7, 14, 21, and 28 to confirm that infection was successful and that transmission of PRRSV to contact controls occurred during the study period. Sera were collected from all Group 2, 3, 4, and 5 pigs on post exposure (PE) Days 0, 2, 7, 14, 21, and 28. Post inoculation Day 5 of Group 1 was equivalent to PE Day 0 of the sentinel groups (Figure 2). Sera were collected from the two negative control pigs (Group 6) at the initiation and completion of each trial.
Sampling of personnel and fomites
In order to assess whether PRRSV could be detected on skin, hair, and mucous membranes of the investigators after the exposure period, swabs of hands, fingers, fingernails, hair, nares, and tonsil were collected(Dacron fiber-tipped plastic applicatorswab; Fisher, Hanover Park, Illinois) before investigators entered the Group 1 room and before they exited from the Group 1 anteroom. The sampling procedure was repeated after performance of each sanitation protocol, before entry into the sentinel rooms. Swab samples were collectedeach day of the exposure period.
Swab samples of personnel were collected using standard operating protocols. The dorsal and ventral surfaces of both hands, the length of each finger, each finger tip, the ventral surface of each fingernail, and the carpal region were sampled. The circumference of the head was swabbed three times in an attempt to sample hair. The nasal passages were swabbed by gently inserting a swab approximately 1.25 cm into each nare. The oropharyngeal region was swabbed in an attempt to sample the tonsil.
Swabs from visibly contaminated areas of boots and coveralls were collected prior to exiting the Group 1 room on each exposure day.
Swabs from personnel and fomites were put into individual sterile tubes (Falcon, Franklin Lakes, New Jersey) containing 2 mL of MEM. Samples were pooled from each anatomical site (hands and fingers, hair, nares, and oropharynx) from each investigatorand the pooled samples were tested for PRRSV by polymerase chain reaction(PCR), virus isolation (VI), and swine bioassay.
Diagnostic analysis
All sera, and swabs collected from the skin, hair, and mucous membranes of investigators and from boots and coveralls, were tested twice by reverse transcriptase PCR for viral nucleic acid (Taqman PCR assay; Perkin-Elmer Applied Biosystems, Foster City, California).14 A sample was considered positive if PRRSV nucleic acid was detected in both tests. Samples positive on only one test were considered suspect.
All sera were tested for PRRSV antibodies by ELISA (IDEXX). An ELISA S:P ratio >=0.4 was considered positive.
All sera, and swabs collected from the skin, hair, and mucous membranes of investigators and from boots and coveralls, were tested for infectious PRRSV by VI using both MARC-145 cells and porcine alveolar macrophages.15 Representative isolates of PRRSV obtained from positive samples were sequenced to compare the percent homology of open reading frame (ORF) 5 region of tested isolates with the same regionof PRRSV strain VR-2332 (the inoculum for Group 1).16
Swabs collected from the skin, hair, and mucous membranes of
investigators and from boots and coveralls were tested by swine
bioassay as described by Swenson
et al.17
Laboratory detection of PRRSV on fomites and personnel
The ability of PCR and VI to detect PRRSV on fomites and personnel
in a laboratory setting was tested in two replicate trials. Five
tenfold dilutions of the PRRSV inoculum were made, and, using
a plastic pipette tip (Redi-Tip; Fisher Scientific, Hanover Park,
Illinois), 0.5 mL of each dilution was dropped onto the coverall
and boot materials used in the study. Each drop was then spread
over a 4 x 4-cm area using a sterile Dacron swab. Each inoculated
site was swabbed immediately
(<5 seconds), 30 minutes, 60 minutes, and 120 minutes after
inoculation, and tested for PRRSV by PCR and VI as described.
In addition, using the same procedure, detectionof PRRSV on the
skin of personnel (hands) was assessed. Prior to inoculation,
hands were washed with hot water and allowed to dry for 1 hour.
The surface of each finger of a hand was then inoculated with
0.5 mL of a specific dilution (one of the five dilutions per finger)
spread over a 4 x 4-cm area using a sterile swab, sampled immediately
(< 5 seconds), and tested for PRRSV by PCR and VI.
Results
Animal samples
All animals were seronegative by ELISA (S:P ratio <0.4) prior to each replicate trial. Negative control pigs (Group 6) were seronegative by ELISA at the end of each trial.
Group 1 (Infected group). In all four trials, PCR and VI showed that 25 of 28 inoculated pigs were infected with PRRSV by PI Day 2, and all 28 were infected by PI Day 5 (Table 1). Transmission of PRRSV from the seven inoculated pigs to the Group 1 in-contact pigs was detected by PCR and VI by PI Day 5, and all three in-contact pigs in each trial were infected by PI Day 7. Nucleic acids in the ORF 5 regions of four representative PRRSV isolates (one isolate per trial) were 99.9% homologous with PRRSV strain VR-2332.
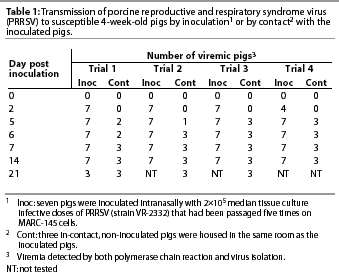
Clinical signs of fever (mean 40°C; range 38.9 to 42°C), anorexia, and lethargy were observed in all inoculated pigs on PI Days 5 to 7 and in all in-contact pigs on PI Days 9 to 12.
By PI Day 14, all seven of the inoculated pigs in each trial were seropositive for PRRSV by ELISA, and at least one of the three in-contact pigs in each trial had seroconverted.
Group 2 (Direct Contact group). Infection with PRRSV occurred in two of the four trials. In Trials 3 and 4, viremia was detected in all six Group 2 pigs (Table 2). Nucleic acids in the ORF 5 regions of two representative isolates (one isolate per trial) were 99.9% homologous with PRRSV strain VR-2332.
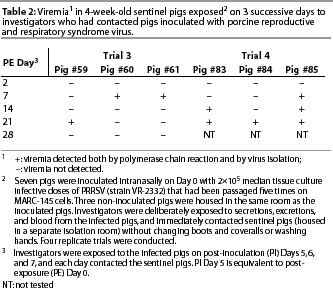
Infected pigs were febrile (mean 40°C; range 39.8 to 40.6°C), anorexic, and lethargic.
In Trial 3, anti-PRRSV antibodies were detected in two of the three pigs by PE Day 14 (Pigs #60 and #61), and in the third pig by Day 21. In Trial 4, anti-PRRSV antibodies were detected in one of the three pigs by PE Day 14 (Pig #85), and in the other two pigs by Day 21.
Groups 3, 4, and 5. In the sentinel pigs in these groups, PRRSV and PRRSV anti-bodies were not detected by any assay at any time during the study.
Personnel and fomite samples
In two of the four trials, PRRSV was isolated from swabs collected from contaminated boots (one investigator) and coveralls (three investigators) immediately after exposure to infected Group 1 pigs.
In one of the four trials, PRRSV nucleic acid was detected on a swab collected from the contaminated hands of an investigator immediately after exposure to the infected pigs in Group 1. Nucleic acid sequencing indicated a high degree of homology (99.9%) with PRRSV strain VR-2332. The PRRSV nucleic acid detected on the swab collected from contaminated hands was positive by swine bioassay.
Neither infectious virus nor nucleic acid was detected by PCR, VI, or swine bioassay on swab samples collected from fomites or personnel after completion of any of the three sanitation protocols.
Laboratory detection of PRRSV on inoculated fomites and personnel
In both trials, all samples from inoculated boots and coveralls were negative for PRRSV by VI at all times after inoculation and at all dilutions of the inoculum. Swabs from inoculated coveralls were PCR-suspect at an inoculum concentration of 104 TCID50 per mL immediately after inoculation, and swabs from inoculated boots and coveralls were PCR-suspect at an inoculum concentration of 105 TCID50 per mL 30 and 60 minutes after inoculation. Other samples were PCR-negative and VI-negative at all times after inoculation and at all dilutions of the inoculum.
Discussion
Our results show that contaminated boots and coveralls may harbor PRRSV for short periods and might transmit PRRSV from infected to susceptible pigs. The same conclusion may be drawn for contaminated hands. While it is unlikely that people could serve as long-term carriers of PRRSV, viral nucleic acid was detected by PCR on the hands immediately after exposure to infected pigs, and was confirmed to be infectiousby swine bioassay. Transmission of the virus was not observed in the sentinel groups where the Danish System, Standard, or Alternative sanitation protocols were used, and PRRSV was not detected in samples from fomites or personnel after completion of these sanitation protocols. Therefore, it appears that under the conditions of this study, the sanitation protocols evaluated were effective in preventing the transmission of PRRSV from infected to susceptible pigs.
This study possessed a number of known strengths and weaknesses. The primary strength of the design was the use of multiple replicates, each with a 3-day exposure period. This may explain the contrast between our results and those of a recently published study which reported no evidence of PRRSV transmission to naive pigs after direct contact with contaminated personneland fomites.10 That study consisted of a single replicate and 1 day of exposure(PI day 7). As details regarding the protocol of exposure were not provided, the exposure methods of the two studies cannot be compared.
Another strength of our study was that multiple diagnostic methods (PCR, VI, and swine bioassay) were used to validate the results, enhancing the accuracy of the data.
The primary limitation of the study was that it did not measure the frequency of the observed events. While all three sanitation protocols were successful in preventing the spread of PRRSV from infected to susceptiblepigs under the conditions of the study, no conclusions can be drawn regarding the differences in their efficacy, as negative results cannot be interpreted. However, if it is assumed that the probability of transmission of PRRSV to the Direct Contact group was 0.5 (50%) on the basis of the outcome of the study (transmission in two of four replicate trials), and the probability of transmission to the Direct Contact group and to the other three groups (Danish System, Standard, and Alternative sanitation protocols) was the same, then the probability of not detecting transmission to each group in the four replicate trials,due to chance alone, would be 6.25% (0.5×0.5×0.5×0.5=0.0625). Since this level of probability may be significant in the case of PRRSV, we must acknowledge the limitations in sensitivity of the diagnostic methods used in the study. In addition, this calculation of probability assumes that the likelihood of transmission was the same for each sanitation protocol group; however, this might not have been true, as contaminated boots and coveralls remained in the Group 2 sentinel room for the 24-hour period following each exposure period.
Other limitations of the study include the testing of a single pathogen, and use of small numbers of pigs housed under ideal environmental conditions. It is unknown whether similar results would be obtained across large populations of pigs housed underfield conditions, or if different pathogens had been tested, eg, transmissible gastroenteritis virus. Therefore, until larger studies can be conducted to measure the repeatability of these data using a statistically sound number of replicates, swine practitioners should not make recommendations to change current biosecurity protocols conducted on clients’ farms, based solely on the results of this study.
Implications
- After exposure to infected pigs, contaminated fomites (boots and coveralls) and hands can transmit PRRSV from infected to susceptible pigs.
- Under the conditions set by this study, three sanitation protocols, commonly employed for the purposes of farm biosecurity in the global swine industry, were effective in preventing transmission of PRRSV from infected to susceptible pigs by fomites and personnel.
- It is strongly recommended that producers and practitioners change boots and coveralls and wash hands between production stages on one-site farms, or between buildings and sites within segregated systems, particularly if there is a difference in PRRSV infection status across pig populations.
Acknowledgement
The authors would like to thank Dr Dave Pyburn for his vision
regarding the importance of this study, and the National Pork
Board and Genetiporc for financial
support.
References – refereed
1. Schoenbaum MA, Freund JD, Beran GW. Survival of pseudorabies virus in the presence of selected diluents and fomites. JAVMA. 1963;198:1393-1397.
3. Sellers RF, Donaldson AI, Herniman KAJ. Inhalation, persistence and dispersal of foot and mouth disease virus by man. J Hygiene. 1970;68:565-573.
4. Wentworth DE, McGregor MW, Macklin MD, Neumann V, Hinshaw VS. Transmission of swine influenza virus to humans after exposure to experimentally infected pigs. J Inf Dis. 1997;175:7-15.
6. Dee SA, Joo HS, Polson DD, Marsh WE. Evaluation of the effects of the nursery depopulation of the profitability of 34 pigs farms. Vet Rec. 1997;140: 498-500.
8. Christopher-Hennings J, Nelson EA, Hines RJ, Nelson JK, Swenson SL, Zimmerman JJ, Chase CL, Yaegar MJ, Benfield DA. Persistence of porcine reproductive and respiratory syndrome virus in serum and semen of adult boars. J Vet Diagn Invest. 1995;7:456-464.
9. Pirtle EP, Beran GW. Stability of porcine reproductive and respiratory syndrome virus in the presence of fomites commonly found on farms. JAVMA. 1996;208:390-392.
11. Otake S, Dee SA, Kurt DR, Joo HS, Deen J, Molitor TW. Transmission of porcine reproductive and respiratory syndrome virus by needles. Vet Rec. In press.
12. Moore C. Biosecurity and minimal disease herds. Vet Clin N Am Food Anim Pract. 1992;8:461-474.
15. Bautista EM, Goyal S, Yoon IJ, Joo HS, Collins J. Comparison of porcine alveolar macrophages and CL 2621 for the detection of porcine reproductive and respiratory syndrome virus (PRRSV) and anti-PRRSV antibody. J Vet Diagn Inv. 1993;5:163-165.
16. Murtaugh MP, Elan MR, Kakach LT. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the porcine reproductive and respiratory syndrome virus. Arch Vir. 1995;40:1451-1460.
17. Swenson SL, Hill HT, Zimmerman JJ. Excretion of porcine reproductive and respiratory syndrome virus after experimentally induced infection in boars. JAVMA. 1994;204:1943-1948.
References – non-refereed
5. Nielsen JP, Fredriksen W. Atrophic rhinitis in pigs caused by a human isolate of toxigenic Pasteurella multocida. Proc IPVS Cong. Lausanne, Switzerland. 1990;75.
13. Snyder ML, Mermer B, Anderson PR, Wensvoort G, Hill HT. Evaluation data for an immunological ELISA for PRRS. Proc 2nd Int Symp PRRSV. Copenhagen, Denmark. 1995:15.
14. Molitor TW, Tune KA, Shin J, Collins, JE, Kapur, V. Application of TaqMan(tm) PCR in the detection of porcine reproductive and respiratory syndrome virus. Proc Allen D. Leman Swine Conf. St Paul, Minnesota. 1997;173-175.