| Original research | Peer reviewed |
Cite as: Greiner LL. Evaluation of the likelihood of detection of porcine epidemic diarrhea virus or porcine delta coronavirus ribonucleic acid in areas within feed mills. J Swine Health Prod. 2016;24(4):198–204.
Also available as a PDF.
SummaryObjective: To compare areas within feed mills to determine the likelihood of presence of either porcine epidemic diarrhea virus (PEDV) or porcine delta coronavirus (PDCoV). Materials and methods: Twenty-four feed mills from various regions in the United States were evaluated. Swab samples (foot pedals of feed delivery trucks, bulk ingredient unloading pits, inside the mixer or pellet coolers, mill office floors, inside feed compartments on feed trucks and incoming bagged-ingredient trucks) were collected daily at each feed mill for up to 5 days. The samples were submitted for polymerase chain reaction testing for PEDV and PDCoV. Results: Of the feed mills tested, 75% were supplying feed to known PEDV-positive herds, and 21% were supplying feed to known PDCoV-positive herds. No samples at any mill tested positive for PEDV ribonucleic acid (RNA), although 5% of the truck foot pedals and 1% of the bulk-ingredient pits tested suspect. Porcine delta coronavirus RNA was found on 3.4% of the foot pedals of the trucks, and 2.2% of the office floors tested suspect. Implications: Porcine delta coronavirus RNA can be detected at different locations around feed mills. Feed mill biosecurity protocols need to be evaluated and maintained to minimize the probability of PEDV and PDCoV RNA presence. | ResumenObjetivo: Comparar áreas dentro de los molinos de alimento para determinar la probabilidad de la presencia del virus de la diarrea epidémica porcina (PEDV por sus siglas en inglés) o del delta coronavirus porcino (PDCoV por sus siglas en inglés). Materiales y métodos: Se evaluaron veinticuatro molinos de alimento de diferentes regiones en los Estados Unidos. En todos los molinos, se recolectaron diariamente durante cinco días, muestras con hisopos (pedales de los camiones de entrega de alimento, pozos de descarga de ingredientes a granel, dentro de las mezcladoras o enfriadores de pellets, pisos de las oficinas del molino, dentro de los compartimientos de alimento en los camiones de alimento, y en los camiones que entregan ingredientes ensacados). Estas muestras fueron enviadas para ser analizadas con la prueba de reacción en cadena de polimerasa en busca de PEDV y PDCoV. Resultados: De los molinos estudiados, 75% estaban supliendo alimento a hatos positivos al PEDV y 21% a hatos positivos al PDCoV. Ninguna muestra en ningún molino resultó positiva al ácido ribonucleico (RNA) de PEDV, sin embargo 5% de los pedales de camión y 1% de los pozos de ingredientes a granel resultaron sospechosos. El RNA del delta coronavirus porcino se encontró en 3.4% de los pedales de los camiones, y 2.2% de los pisos de oficina resultaron sospechosos. Implicaciones: El RNA del delta coronavirus porcino puede ser detectado en diferentes localizaciones alrededor de los molinos de alimento. Los protocolos de bioseguridad de los molinos de alimento deben ser evaluados y mantenidos para minimizar la probabilidad de la presencia del RNA del PEDV y PDCoV. | ResuméObjectif: Comparer des sites à l’intérieur de meuneries afin de déterminer la présence possible du virus de la diarrhée épidémique porcine (VDEP) ou du coronavirus delta porcin (CoVDP). Matériels et méthodes: Vingt-quatre meuneries de différentes régions aux États-Unis ont été évaluées. Des écouvillonnages (pédales des camions de livraison de moulée, fosses de déchargement des ingrédients en vrac, intérieur du mélangeur ou refroidisseurs des granules, planchers des bureaux de la meunerie, intérieur des compartiments de moulée des camions de nourriture, camions amenant les ingrédients en sac) ont été faits quotidiennement jusqu’à 5 jours à chaque meunerie. Les échantillons ont été analysés par réaction d’amplification en chaîne par la polymérase pour détecter VDEP et CoVDP. Résultats: Des différentes meuneries testées, 75% fournissaient de la moulée à des troupeaux connus pour être positifs au VDEP et 21% fournissaient de la moulée à des troupeaux connus pour être positifs au CoVDP. Aucun échantillon provenant des meuneries testées ne s’est avéré positif pour la présence d’acide ribonucléique (ARN) du VDEP, bien que les tests pour 5% des pédales des camions et 1% des fosses d’ingrédients en vrac fussent suspects. L’ARN du CoVDP fut trouvé sur 3,4% des pédales des camions, et 2,2% des planchers des bureaux donnaient un résultat suspect. Implications: L’ARN du CoVDP peut être détecté dans différents sites autour des meuneries. Les protocoles de biosécurité des meuneries doivent être évalués et maintenus afin de minimiser la probabilité de la présence d’ARN du VDEP et du CoVDP. |
Keywords: swine, feed mill, porcine epidemic diarrhea virus, sampling, probability assessment, PEDV
Search the AASV web site
for pages with similar keywords.
Received: August 7, 2015
Accepted: April 7, 2016
Porcine epidemic diarrhea virus (PEDV) was first detected in the United States in the spring of 2013.1 Porcine delta coronavirus (PDCoV) was first reported in the United States early in 2014; however, retrospective analysis of stored samples demonstrated that the virus was present in the fall of 2013.2 In the retrospective analysis of PDCoV presence in the United States, a questionnaire was used to identify potential sources of introduction of the virus into the farms. Areas reviewed in the questionnaire included vehicles on the farm, as well as drivers and sources of feed.2 With both feed trucks and staff returning from swine farms to the feed mill and incoming ingredient trucks delivering feedstuffs to the feed mill, an assessment was needed to determine the likelihood that, on any given day, viral ribonucleic acid (RNA) might be present in various areas of the mill and mill-associated fomites (eg, transport vehicles, footwear). Therefore, the goal of this study was to investigate the likelihood of detecting either PEDV or PDCoV RNA in selected areas within commercial feed mills.
Materials and methods
Twenty-four feed mills from eight states in the United States were evaluated in this study. The study was conducted from March 17 to September 1 of 2014. Six of the feed mills were providing feed to herds negative for PEDV. Feed mills were considered to be supplying feed to positive herds if one or more client farms were known to be positive for PEDV, PDCoV, or both viruses. The herd status information was reported from either the veterinarians that were involved with the feed mills or the feed mill managers to the individuals collecting the swab samples. The number of sites to which each mill delivered feed ranged from one to 173, with the median 32.5 sites. For up to 5 days, samples were collected daily at each feed mill to estimate the probability of a feed mill testing positive for PEDV or PDCoV. Sample areas included mill office floor, bulk ingredient unloading pit grate, incoming bagged-ingredient truck (inside of truck near site of off-loading), interior of either the mixer or pellet cooler, interior of one feed compartment on a feed truck, and both foot pedals of a feed delivery truck.
In brief, sample kits were prepared before the collection period at the mill, each including latex gloves; 50-mL tube containing 5 mL of sterile phosphate buffered saline (PBS); sealable plastic bag containing 25 mL of sterile PBS; and 10.16 cm × 10.16 cm sterile gauze. The sterile gauze was placed in the tube with the PBS and remained there until used. The collector changed gloves between samples. The gauze was removed from the 50-mL tube and wiped over the entire sample area (approximately 0.09 m2). After each collection, the soiled gauze was placed in the sealable plastic bag and squeezed to express the fluid. Fluid was then drained from the bag into its original 50-mL tube and labeled accordingly (location within mill, mill identity, date). The tubes were then placed on ice in a cooler for transport. Samples were kept frozen in a -20˚C freezer for the week prior to submission. Samples were tested for PEDV and PDCoV RNA via polymerase chain reaction (PCR) at the University of Minnesota (19 mills), Iowa State University (three mills), South Dakota State University (one mill), and a privately-owned laboratory (one mill). Each laboratory reported its respective cutoffs for a positive, negative, and suspect cycle threshold (Ct) value (University of Minnesota: < 35 positive, 35 to ≤ 40 suspect, > 40 negative; Iowa State University: < 36 positive, ≥ 36 negative; South Dakota State University: < 38 positive, ≥ 38 negative; private laboratory: ≤ 40 positive, > 40 negative), and those cutoffs were used in the analysis. All samples collected were tested for PEDV, and for 19 of the 24 mills, samples were also tested for PDCoV.
Polymerase chain reaction results were identified as positive, suspect, or negative on the basis of the respective diagnostic laboratory-designated cutoffs. Feed mills were assigned a letter identity for anonymity during analysis and reporting.
Data were analyzed to determine probability of PEDV or PDCoV RNA particle presence using the Bayesian model. Each sample was considered an experimental unit and was blocked both by location within the mill and health status of the site serviced by the mill. In addition, data was weighted on the basis of the percentage of mills that were supplying feed to either negative-status or positive-status herds. Data were summarized to indicate probabilities of detection in samples for each area being tested within the mills. In addition, probabilities of detection were calculated for the subset of mills that were known to be servicing positive herds.
Results
Seventy-five percent (18) of the feed mills tested were providing feed to PEDV-positive herds and 21% (five) of the feed mills were providing feed to PDCoV-positive herds. One third (eight) of the feed mills reported “unknown” concerning the PDCoV status of the herds that they were supplying with feed.
Raw means from the total samples collected per location within the mills for both PEDV and PDCoV are presented in Tables 1 and 2, respectively. No samples tested positive for PEDV. However, 5% of the truck foot pedals and 1% of the bulk ingredient pits tested suspect for PEDV. Porcine delta coronavirus RNA was found on 3.4% of the foot pedals of the trucks, and 2.2% of the office floors tested suspect. One mill that was currently not known to be supplying feed to PDCoV-positive herds did have a PDCoV suspect result on a sample from the office floor.
Table 1: Proportions (%) of feed mill samples positive for porcine epidemic diarrhea virus (PEDV) ribonucleic acid by sampling site*
| Office floor | Bulk ingredient pit | Ingredient delivery truck | Mixer/cooler | Feed truck compartment | Foot pedal | |
|---|---|---|---|---|---|---|
| No. of samples | ||||||
| Total | 100 | 100 | 74 | 99 | 100 | 100 |
| Polymerase chain reaction results (%) | ||||||
| Positive | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Suspect | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 5.0 |
| Negative | 100.0 | 99.0 | 100.0 | 100.0 | 99.0 | 95.0 |
Table 2: Proportions (%) of feed mill samples positive for porcine delta coronavirus ribonucleic acid by sampling site*
| Office floor | Bulk ingredient pit | Ingredient delivery truck | Mixer/cooler | Feed truck compartment | Foot pedal | |
|---|---|---|---|---|---|---|
| No. of samples | ||||||
| Total | 100 | 100 | 74 | 99 | 100 | 100 |
| Polymerase chain reaction results (%) | ||||||
| Positive | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.4 |
| Suspect | 2.2 | 0.0 | 0.0 | 0.0 | 0.0 | 1.1 |
| Negative | 97.8 | 100.0 | 100.0 | 100.0 | 100.0 | 95.5 |
Tables 3 and 4 demonstrate the probability of negative results on the basis of the perceived health status of the herds to which the mills were supplying feed. In short, the areas that tested either positive or suspect had a lower probability of testing negative over time. Overall, mills that were supplying feed to at least one positive herd had a lower probability of testing negative.
Table 3: Average probability of negative porcine epidemic diarrhea virus (PEDV) polymerase chain reaction (PCR) test results on the basis of location and the PEDV status (positive or negative) of the herds being supplied with feed from the mill*
| Average (over five samples) probability of negative PCR test results | |||||||
|---|---|---|---|---|---|---|---|
| Office floor | Bulk ingredient pit | Ingredient truck | Mixer/cooler | Truck compartment | Foot pedal | ||
| Mills feeding PEDV-positive pigs | |||||||
| Test ~+† | 1.000 | 0.983 | 1.000 | 1.000 | 0.986 | 0.931 | |
| Test -‡ | 1.000 | 0.983 | 1.000 | 1.000 | 0.986 | 0.931 | |
| Mills feeding PEDV-negative pigs | |||||||
| Test ~+ | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| Test - | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| Total mills | |||||||
| Test ~+ | 1.000 | 0.989 | 1.000 | 1.000 | 0.990 | 0.951 | |
| Test - | 1.000 | 0.989 | 1.000 | 1.000 | 0.990 | 0.951 | |
* Study described in Table 1.
† Test ~+: Either a positive or suspect result.
‡ Test -: Negative result.
Table 4: Average probability of negative porcine delta coronavirus (PDCoV) polymerase chain reaction (PCR) test results on the basis of location and the PDCoV status (positive or negative) of the herds being supplied with feed from the mill*
| Average (over five samples) probability of negative PCR test results | ||||||
|---|---|---|---|---|---|---|
| Office floor | Bulk ingredient pit | Ingredient truck | Mixer/cooler | Truck compartment | Foot pedal | |
| Mills supplying feed for PDCoV-positive pigs | ||||||
| Test +† | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 0.840 |
| Test -‡ | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 0.840 |
| Mills supplying feed for pigs of unknown PDCoV status§ | ||||||
| Test + | 0.967 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| Test - | 0.967 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| Mills supplying feed for PDCoV-negative pigs | ||||||
| Test + | 0.967 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| Test - | 0.967 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| Total mills | ||||||
| Test + | 0.982 | 1.000 | 1.000 | 1.000 | 1.000 | 0.923 |
| Test - | 0.982 | 1.000 | 1.000 | 1.000 | 1.000 | 0.923 |
* Study described in Table 1.
† Test +: Positive result
‡ Test -: Negative result
§ PDCoV status of herds being supplied with feed from the mills was not known at the time of the study, as PDCoV PCR was not a diagnostic test performed at that time for those sites.
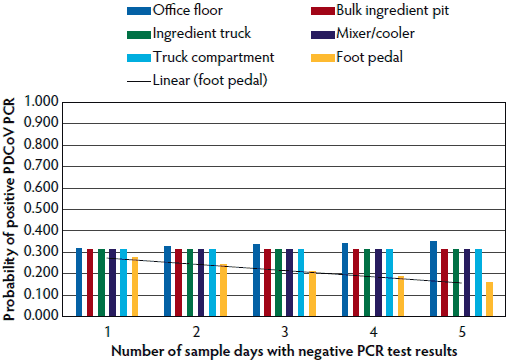
Tables 5 and 6 further break down the probability of either PEDV or PDCoV positive or suspect results at the various locations tested within mills in relation to the perceived health status of the herds they supplied. As the number of sampling days increased, the probability of a positive or suspect result increased independently for the foot pedals, bulk ingredient pit, and the inside of the feed truck compartment for PEDV, and for the foot pedals and office floor for PDCoV.
Table 5: Probability of having a porcine epidemic diarrhea virus (PEDV) positive or suspect polymerase chain reaction (PCR) test result on the basis of the number of days tested and the PEDV status (positive or negative) of the herds being supplied with feed from the mill*
| Probability of any positive PCR test results by sample days | |||||||
|---|---|---|---|---|---|---|---|
| Sample days | Office floor | Bulk ingredient pit | Ingredient truck | Mixer/cooler | Truck compartment | Foot pedal | |
| Mills supplying feed for PEDV-positive pigs | |||||||
| Test ~+† | 1 | 0.000 | 0.017 | 0.000 | 0.000 | 0.014 | 0.069 |
| 2 | 0.000 | 0.033 | 0.000 | 0.000 | 0.028 | 0.133 | |
| 3 | 0.000 | 0.049 | 0.000 | 0.000 | 0.042 | 0.193 | |
| 4 | 0.000 | 0.065 | 0.000 | 0.000 | 0.056 | 0.248 | |
| 5 | 0.000 | 0.081 | 0.000 | 0.000 | 0.069 | 0.300 | |
| Mills supplying feed for PEDV-negative pigs | |||||||
| Test ~+† | 1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 4 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 5 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
* Study described in Table 1.
† Test ~+ : Either a positive or suspect result.
Table 6: Probability of having a porcine delta coronavirus (PDCoV) positive or suspect polymerase chain reaction (PCR) test result on the basis of the number of days tested and the PDCoV status (positive or negative) of the herds being supplied with feed from the mill*
| Probability of any positive PCR test results by sample days | |||||||
|---|---|---|---|---|---|---|---|
| Sample days | Office floor | Bulk ingredient pit | Ingredient truck | Mixer/cooler | Truck compartment | Foot pedal | |
| Mills feeding PDCoV-positive pigs | |||||||
| Test+ | 1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.160 |
| 2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.294 | |
| 3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.407 | |
| 4 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.502 | |
| 5 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.582 | |
| Mills feeding pigs of unknown PDCoV status† | |||||||
| Test+ | 1 | 0.033 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 2 | 0.066 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 3 | 0.097 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 4 | 0.127 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 5 | 0.156 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Mills feeding PDCoV- negative pigs | |||||||
| Test+ | 1 | 0.033 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 2 | 0.066 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 3 | 0.097 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 4 | 0.127 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 5 | 0.156 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
* Study described in Table 1.
† PDCoV status of herds being supplied with feed from the mills was not known at the time of the study, as PDCoV PCR was not a diagnostic test performed at that time for those sites.
Figures 1 and 2 demonstrate the probabilities of a mill testing positive on the basis of the number of days it has tested negative, with the percentage of mills supplying feed to positive herds included in the analysis.
Figure 1: Probability of detecting porcine epidemic diarrhea virus (PEDV) ribonucleic acid particles, determined by the number of sequential negative results on polymerase chain reaction (PCR) testing. Study described in Table 1.
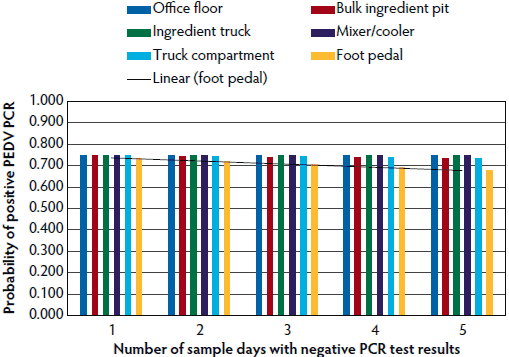
Figure 2: Probability of detecting porcine delta corona virus (PDCoV) ribonucleic acid particles, determined by the number of sequential negative results on polymerase chain reaction (PCR) testing. Study described in Table 1.
Discussion
The areas of the feed mill and delivery trucks were purposively selected for testing due to the perceived potential for either cross-contamination by foot or truck traffic in the mill or farm or as an indirect measurement of the feed as a potential vector. For example, testing the foot pedals and office floor should reflect the potential for feed-mill staff or visitors to transfer contaminants from the farm to the mill or vice versa. Testing the incoming ingredient pit and delivery truck were chosen to evaluate the potential for the viruses to be transmitted into the feed mill with incoming ingredients. Finally, evaluation of the mixer and the inside of the feed truck allowed for assessment of the potential for the final feed product to be a source of viral transmission.
Other areas of the mill, such as the micronutrient intake point and ingredient bags or totes, could have also been tested. Recent data3 demonstrated that PEDV in the feed would be widespread in a feed mill after manufacturing feed that contained PEDV. Therefore, testing the mixer and the inside of the feed delivery truck would provide information as to whether or not specific ingredients or the mixer intake points could serve as a source of viral introduction.
Although no samples tested positive for PEDV RNA, suspect results were seen in samples from the foot pedals of feed delivery trucks and mill office floors. However, PDCoV PCR-positive samples were found on feed truck foot pedals, and several PCR-suspect results were found in samples from the office floors of the tested feed mills. Mills currently feeding PEDV-positive or PDCoV-positive pigs had a higher chance of having a positive or suspect PCR test. However, the percentage of feed mills supplying feed to at least one PEDV-positive herd was higher than the percentage of those supplying at least one PDCoV-positive herd. As expected with sampling for low frequency events, repeated testing over time led to more mills yielding positive results. For example, after 1 day of negative test results on foot pedals, there was a one in 1.4 chance that the foot pedals would test positive for PEDV with additional days of testing.
One mill that was not supplying feed to any known PDCoV-positive herds at the time of testing did have a PDCoV-suspect result on a sample from the office floor, indicating that the health statuses of the herds being serviced by the mill are not the only potential sources that could lead to a positive or suspect PCR test. The status of the herd serviced by this mill was confirmed both before and after the study by the herd veterinarian.
Although this study did not determine if the suspect or positive PCR samples would result in creating a clinical disease in an animal, data from Thomas et al4 demonstrates that 25% of the time, when PCR samples have a Ct value of > 45, samples may be infectious. Therefore, on the basis of this information, both positive and suspect samples in this study appear to have the potential to be infectious. Furthermore, the data from the current feed-mill study demonstrated that positive samples can be found in feed mill and delivery trucks, indicating that PEDV and PDCoV control practices should be in place at the feed mill.
Implications
• Under the conditions of this study, porcine delta coronavirus RNA can be detected at different locations around feed mills.
• Feed mill biosecurity protocols need to be evaluated and maintained to minimize the probability of PEDV and PDCoV RNA presence.
Acknowledgements
This study was fully funded by the National Pork Board. The author would like to thank Dr Brent Frederick and Dr Chad Hastad as co-collaborators on the grant, all the individuals who helped gather the samples at the mills, and Dr Kurt Brattain for assisting in the data analysis.
Disclaimer
Scientific manuscripts published in the Journal of Swine Health and Production are peer reviewed. However, information on medications, feed, and management techniques may be specific to the research or commercial situation presented in the manuscript. It is the responsibility of the reader to use information responsibly and in accordance with the rules and regulations governing research or the practice of veterinary medicine in their country or region.
References
1. Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. 2013;25:649–654. doi: 10.1177/1040638713501675.
2. McCluskey BJ, Haley C, Rovira A, Main R, Zhang Y, Barder S. Retrospective testing and case series study of porcine delta coronavirus in US swine herds. Prev Vet Med. 2016;123:185–191. doi: 10.1016/j.prevetmed.2015.10.018.
*3. Schumacher LL, Cochrane RA, Evans CE, Kalivoda JR, Woodworth JC, Stark CR, Jones CK, Main RG, Zhang J, Dritz SS, Gauger PC. Evaluating the effect of manufacturing porcine epidemic diarrhea virus (PEDV)-contaminated feed on subsequent feed mill environmental surface contamination. Kansas Agricultural Experiment Station Research Reports. 2015;1:7.
4. Thomas JT, Chen Q, Gauger PC, Gimenez-Lirola LG, Sinha A, Harmon KM, Madson DM, Burrough ER, Magstadt DR, Salzbrenner HM, Welch MW, Yoon K, Zimmerman JJ, Zhang J. Effect of porcine epidemic diarrhea virus infectious doses on infection outcomes in naive conventional neonatal and weaned pigs. PLoS One. 2015;10(10):e0139266. doi:10.1371/journal.pone.0139266.
* Non-refereed reference.
